Gene synthesis method
A gene synthesis and gene technology, applied in the field of gene synthesis, can solve the problems of writing genes in cumbersome experimental steps, limiting throughput and per capita production capacity, material resources and time consumption, etc., achieving the effect of clear background, increasing per capita production capacity, and ensuring accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0033] The embodiment gene synthesis method of the present invention, described method comprises:
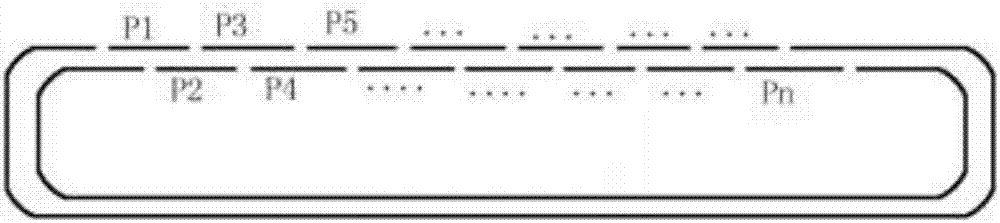
[0034] (1) Divide the gene sequence to be synthesized into the first continuous fragment of 55-100bp length, and design the first continuous fragment as a forward primer; use the gene sequence at a length of 15-20bp staggered from the first continuous fragment as the first continuous fragment Two continuous fragments, designing continuous reverse primers on the reverse complementary sequence of the second continuous fragment;
[0035] (2) mixing the forward primer and the reverse primer to obtain a primer mixture;
[0036] (3) boiling and annealing the primer mixture to obtain double-stranded DNA fragments containing multiple nicks;
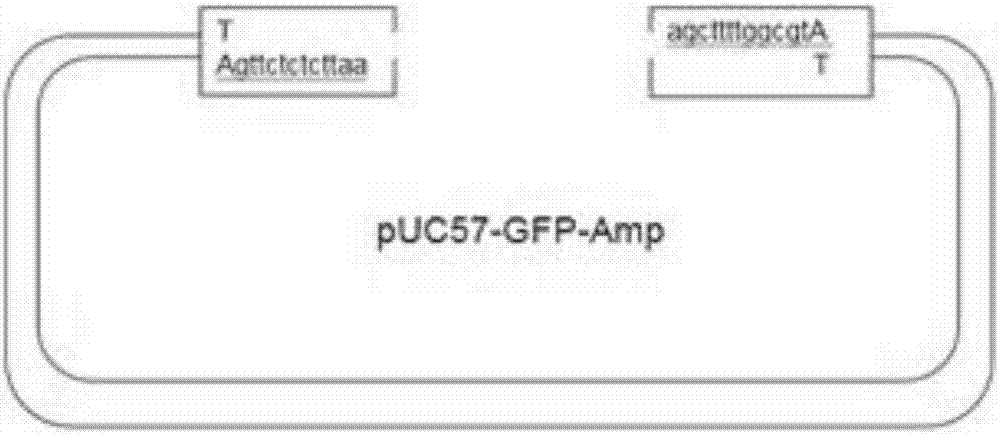
[0037] (4) The synthetic double-stranded DNA fragment was connected to the pUC57-GFP-Amp carrier, the sequence of the carrier was as shown in Sequence Table 1, transformed into Escherichia coli cells, reverse screening of non-fluorescent bacterium...
Embodiment 1
[0048] The preparation method of pUC57-GFP-Amp vector includes:
[0049] 1. A pUC57 vector is provided, and the SapI site on the pUC57 vector is mutagenized to remove the SapI site. Then, the MCS (Multiple Cloning Site) was modified to retain only the EcoRI and HindIII sites at both ends, and a sequence was designed in front of the EcoRI site and behind the HindIII site respectively, so that a relatively long sticky DNA was obtained after one treatment with T4 DNA polymerase. Sexual end effect. A GFP ORF (open reading frame) was filled between the EcoRI and HindIII sites, and the vector was named pUC57-GFP-Amp.
[0050] The specific steps are as follows:
[0051] 1.1 Mutagenesis of the SapI site: design primers (underlined bases are mutated bases)
[0052] pUC57-SapImut-F:5'GCGTATTGGGCGC A CTTCCGCTTCCTCGCTCACTGACTC3'
[0053] pUC57-SapImut-R:5'GCGAGGAAGCGGAAG T GCGCCCAATACGCAAAC 3'
[0054] The pUC57-Amp vector was used as a template to amplify, and the amplified produc...
Embodiment 2
[0094] The preparation method of the pUC57-GFP-Amp vector is similar to that of Example 1 and will not be repeated here.
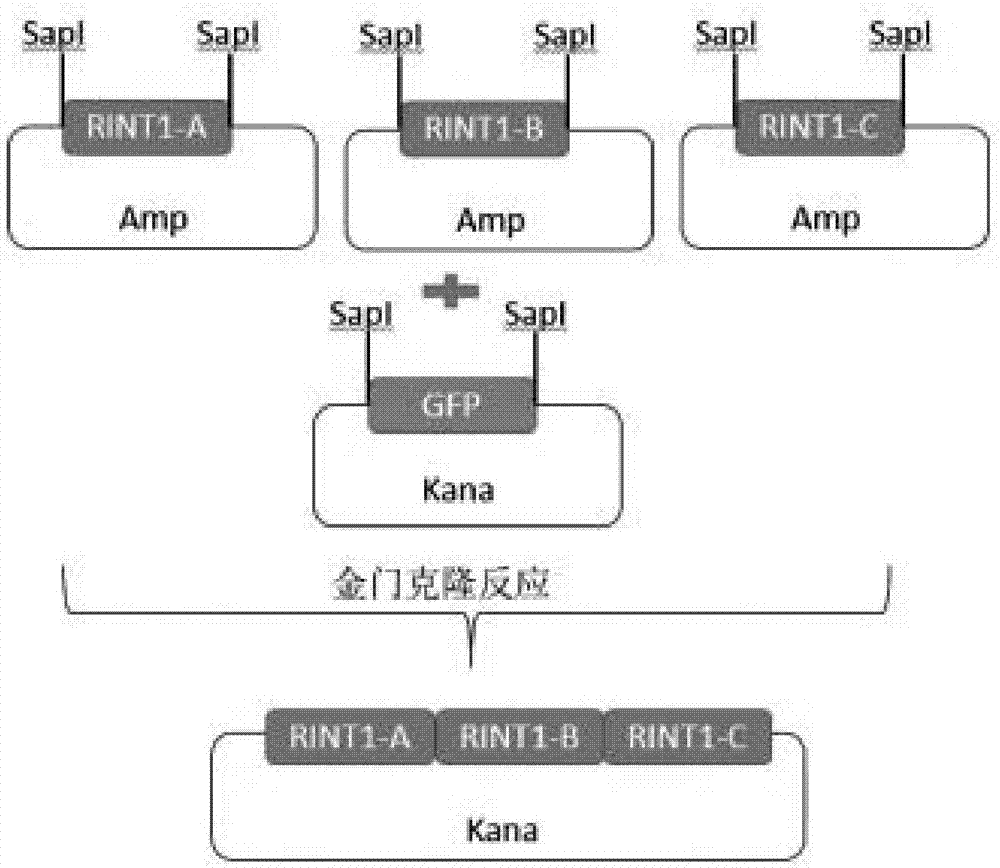
[0095] 1. Gene grouping (the underline is the annealing sequence with the vector, the wavy line is the BbsI site, and the italic part is the Golden Gate cloning reaction site)
[0096] 16128A(747bp)
[0097] caagagagaatt c ccatggcaacaggaactgagaaaaagcaccaagaaaagctttatatccctaagctaaaggttcatcagatcaagctagagctcttttatgaggccggtcatggacaggtggcctttgataacctttctttgagagaggctggtgataagccaagtgatgacatcaaggttgcttcacatcacttagaggagcaaatcgtcctgcctttaaataagcactacctcatggaaatggctgactatcactatcagattgcagcggagtccagcaacattgtgcgcgtggaaaatggtcttttgattcctttgtctcacgggaaaacgcttttagaggtgttggatcaagaggggcaaagagtagcaacagtgccggttgagattttagcagcagaggaccctcaaacaacctccttgatcacaaagtggtgtgaggtgattttgggggctgaaaactttgaccgatcaagtccagctatggtggcattaaaccaaaaactggacgacagtgtgagcaaaaatctagttcaaatatgtatccgctcatgagacaataaccctgataaatgcttcaataatattgaaaaaggaagagtcctgaggcggaaagaaccagctgtggaatgtgtgtcagttagggtgtgga...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com