Method for performing secretory expression of glucose oxidase based on optimization of metabolic engineering, recombinant bacterium and application thereof
A technology of glucose oxidase and glucose, applied in the direction of microorganism-based methods, genetic engineering, oxidoreductase, etc., can solve the problems of unsatisfactory yield and enzyme activity, GOD inactivity, difficulty in separating and purifying intracellular enzymes, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0120] Embodiment 1, the construction of recombinant bacterial strain
[0121] 1. Construction of recombinant bacteria G / GODM
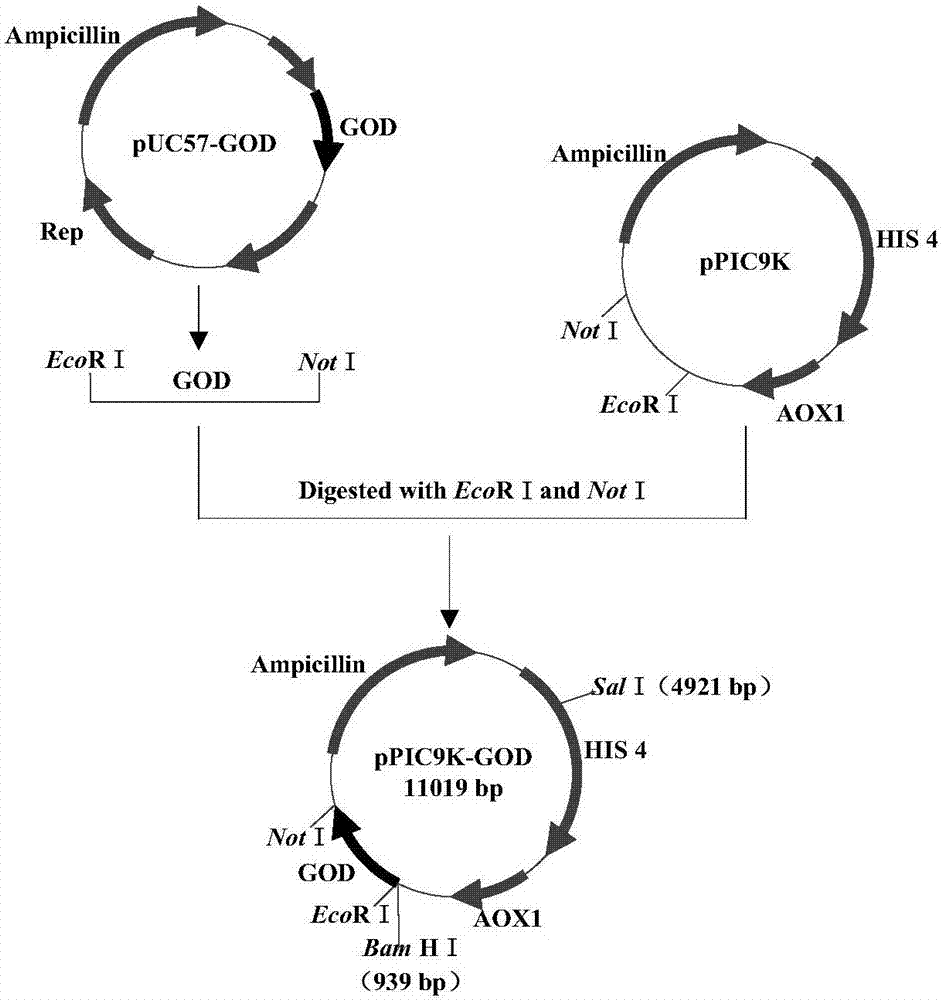
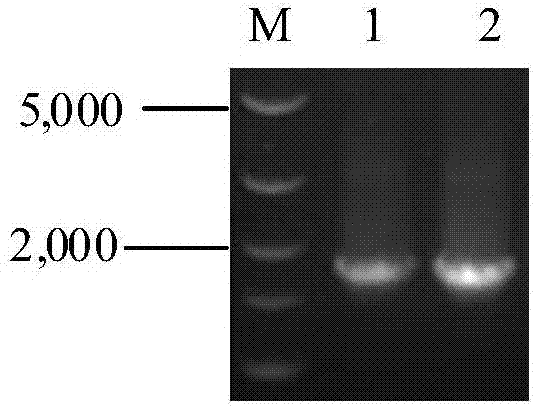
[0122] In order to realize the secreted expression of glucose oxidase in Pichia pastoris, the inventors selected the pPIC9K vector. Using pUC57-GOD as a template, by PCR, introduce EcoR 1 at the 5' end of the GOD sequence, introduce Not 1 at the 3' end, digest with enzymes, and connect with pPIC9K cut with the same enzyme to construct the GOD secretion expression vector pPIC9K-GOD, construction flow chart Such as figure 1 . More specifically, using the plasmid pUC57-GOD (the GOD nucleic acid sequence is connected to pUC57) as a template, using primers GODF and GODR, the target fragment GOD (1749bp, see figure 2 ), introduce EcoR Ⅰ and Not 1 two enzyme cutting sites at the same time, use restriction endonucleases to linearize the amplified product and pPIC9K vector respectively, then perform ligation reaction, and finally, transform into E. coli DH...
Embodiment 3
[0130] Embodiment 3, the performance investigation of recombinant bacteria
[0131] Pick the verified transformants G / GMS3 and G / GMM1, and inoculate them into a test tube containing 3mL of YPG medium. After culturing for about 18 hours, transfer them to a 250mL shake flask containing 25mL of BMGY medium, and enrich the culture until OD 600 4 to 6, collect the bacteria, inoculate into 500mL shake flask containing 50mL BMMY medium for culture, initial OD 600=1, add 1% methanol solution every 24h, and take samples to measure OD at the same time 600 , to investigate the effects of co-expression of zwf1, sol3, gdh3, mdh1 on the growth of recombinant bacteria, changes in enzyme activity, etc.
[0132] 1. Cell growth
[0133] Depend on Figure 9 It can be seen that the growth trend of the recombinant bacteria G / GMZ1, G / GMS3, G / GMG3, and G / GMM1 is very close to that of the original bacteria G / GODM, and the average specific growth rate μ is 0.039h -1 left and right, indicating tha...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com