Quality control product for detecting fragmented DNA (Deoxyribonucleic Acid) mutation and preparation method thereof
A technology of fragmentation and quality control products, which is applied in the field of clinical testing and can solve problems such as inability to evaluate the accuracy of external quality
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0031] A method for preparing a quality control product for detecting fragmented DNA mutations, comprising the steps of:
[0032] S1: mutant DNA fragment;
[0033] Wherein, the specific steps for preparing mutant DNA fragments in S1 can be obtained in the following three ways;
[0034] Using the mutant genomic DNA as a template, the mutant DNA fragments are obtained through primer amplification.
[0035] Or use mutant genomic DNA as a template, amplify with primers to obtain mutant DNA fragment precursors, and connect the mutant DNA fragment precursors to plasmids to obtain mutant plasmids.
[0036] Amplify or digest with primers to obtain mutant DNA fragments.
[0037] Or use the DNA whose target gene locus is the wild-type locus as a template, and amplify with primers to obtain wild-type DNA fragments.
[0038] The wild-type DNA fragment was ligated with the plasmid to construct the wild-type plasmid.
[0039] Then use the wild-type plasmid as a template and amplify with...
Embodiment 1
[0066] Main reagents and materials
[0067] KOD Plus Mutagenesis Kit was purchased from Toyobo (Shanghai) Biotechnology Co., Ltd.
[0068] Primers used in PCR were purchased from Sangon Bioengineering (Shanghai) Co., Ltd.
[0069] Competent Escherichia coli DH5α, Universal DNA purification and recovery kit, and endotoxin-free plasmid mid-scale extraction kit were purchased from Tiangen Biochemical (Beijing) Co., Ltd.
[0070] The pMD18-T vector plasmid was purchased from Bao Biological Engineering (Dalian) Co., Ltd.
[0071] Tissue genome extraction reagent QIAamp DNA FFPE Tissue Kit and whole blood genome extraction reagent QIAampDNA blood Mini Kit were purchased from QIAGEN China (shanghai) Co., Ltd.
[0072] Plasma cell-free DNA extraction and purification kit (adsorption column method) and human EGFR gene mutation quantitative detection kit (fluorescent PCR method) were purchased from Gnosper Biotechnology Nantong Co., Ltd.
Embodiment 2
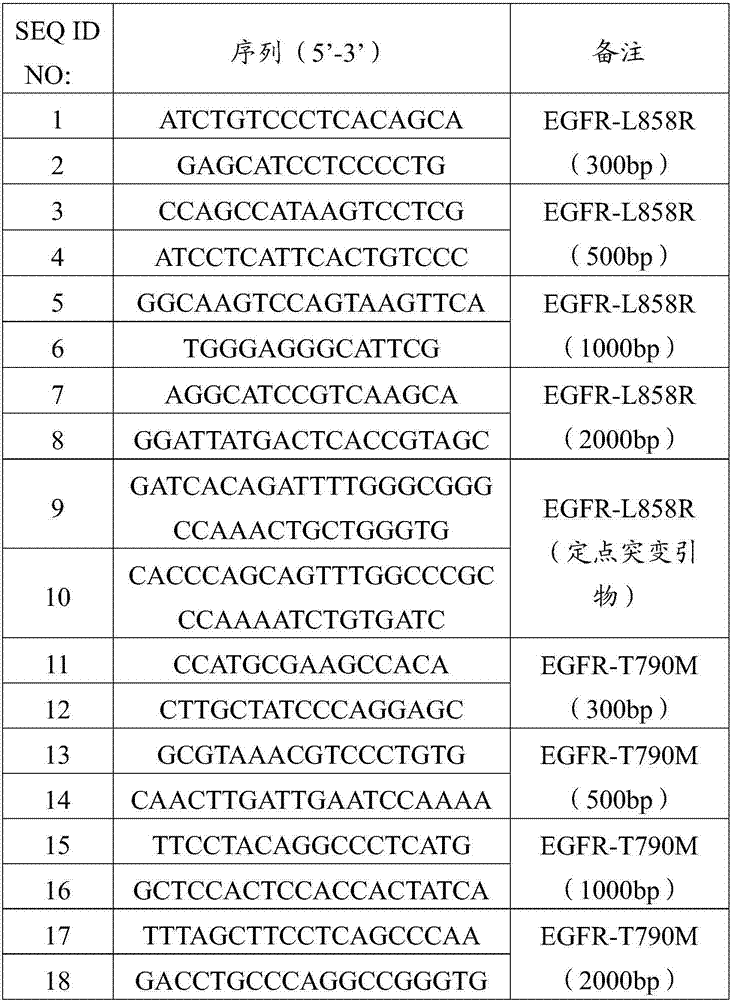
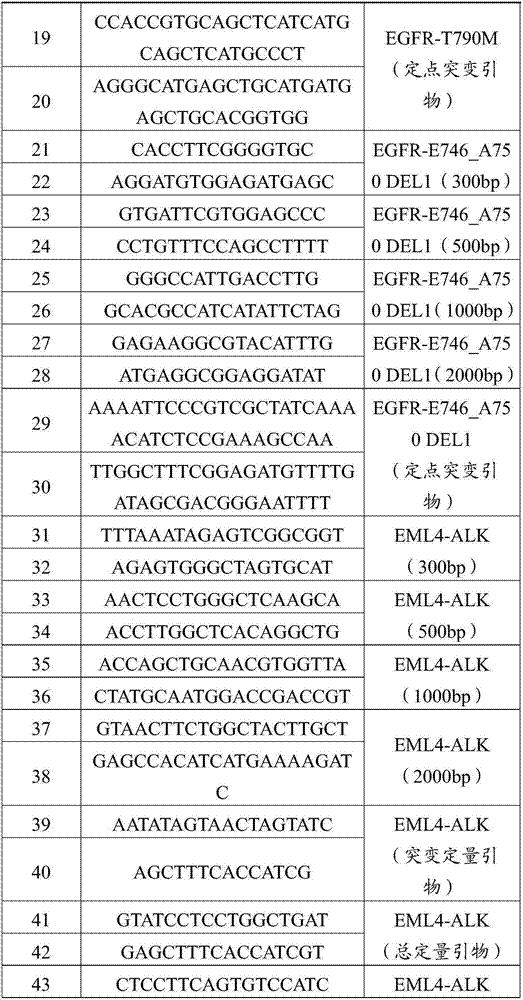
[0093] The T790M wild-type DNA of exon 20 of the human EGFR gene was extracted with a whole blood genome extraction kit (QIAamp DNA blood Mini Kit). The experimental method is basically the same as in Example 1, except that primers (SEQ ID NO: 17 to SEQ ID NO: 18 in Table 1) are used to amplify and amplify a wild-type DNA fragment with a length of 2000 bp to construct a wild-type plasmid . The primer sequences for site-directed mutagenesis are selected from SEQ ID NO:19 to SEQ ID NO:20 in Table 1; the primer sequences required for the construction of mutant DNA fragments are selected from SEQ ID NO:11 to SEQ ID NO:18 in Table 1.
[0094] The T790M mutant fragmented DNA sample of exon 20 of the human EGFR gene was mixed with the extracted healthy human plasma free DNA. Among them, the T790M mutant fragmented DNA sample of exon 20 of the human EGFR gene accounts for 0.1% of the wild-type DNA copy number in the extracted healthy human plasma free DNA. Get the quality control D1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com