pMG36e pgsA gp85 recombinant plasmid-containing genetically engineered bacterium
A pmg36e-pgsa-gp85, genetically engineered bacteria technology, applied in the field of biology, can solve the problems of complex antigen structure, difficult attenuated vaccines, destroying antibodies, etc., and achieve the effect of increasing average daily weight gain, reducing risks, and improving ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0063] Embodiment 1 Amplification of gp85 gene and pgsA gene
[0064] (1) Design specific primers:
[0065] The PCR primer 5'-C was used in the PCR amplification of the target fragment pgsA GAGCTC GCGAACTGAGCTTTCATGAAAAG-3', its nucleotide sequence is shown in SEQ ID NO.1; 5'-CTAG TCTAGA CTATGATCAATATCAAACGTCA-3', the nucleotide sequence of which is shown in SEQ ID NO.2;
[0066] The PCR primer 5'-TCA was used in the PCR amplification of the target fragment gp85 TCTAGA GGGAGTTCATCTGTTG-3', its nucleotide sequence is shown in SEQ ID NO.3 and 5'-TCC AAGCTT ATTAGCGCCTGCTAC-3', the nucleotide sequence of which is shown in SEQ ID NO.4;
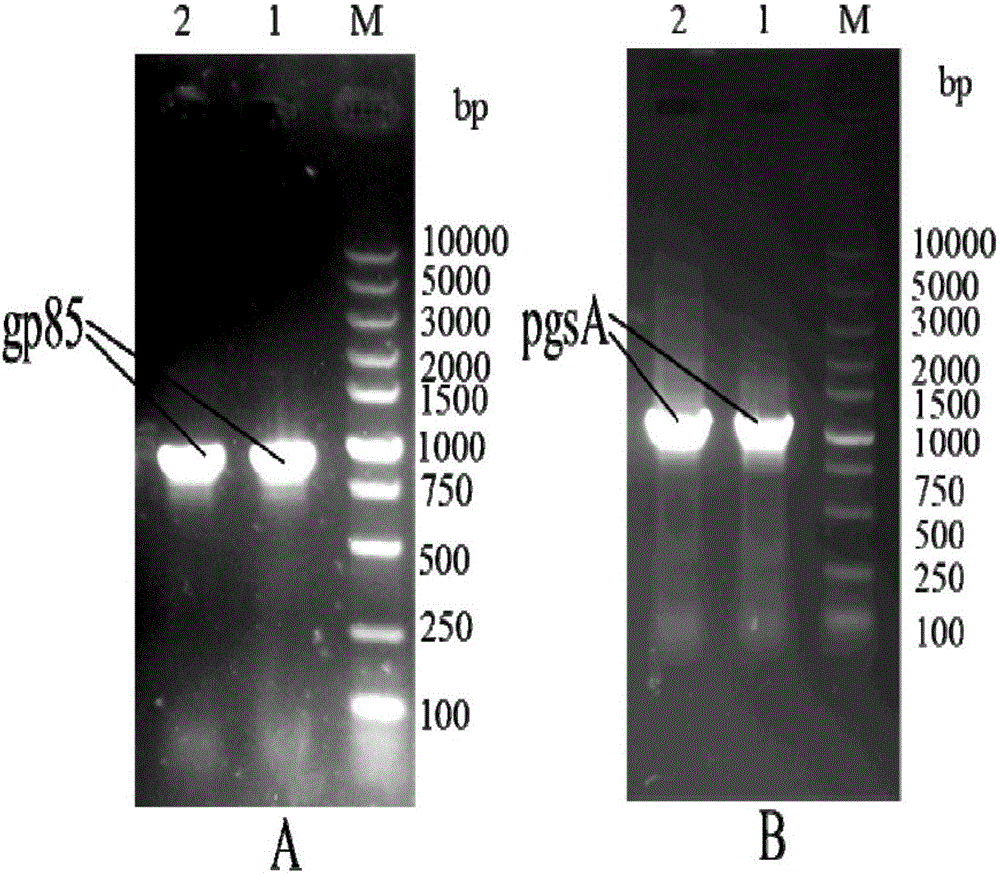
[0067] Utilize the above primers to amplify the gp85 envelope glycoprotein gene from the J subgroup avian leukosis virus env gene; amplify the pgsA anchor protein gene from the pgsA, pgsB and pgsC genes in the whole gene of Bacillus subtilis; the results are as follows figure 2 shown;
[0068]PCR amplification system and conditions of gp8...
Embodiment 2
[0075] The PCR products of gp85 and pgsA gene obtained in step (1) are respectively connected to two separate pMD18-T vectors;
[0076] The pMD18-T vector was ligated with the purified and recovered two target gene fragments respectively. The ligation system was 0.5 μl of pMD18-T vector, 5.0 μl of Solution Ⅰ, and 4.5 μl of purified PCR product. Mix the ligation product evenly, centrifuge slightly to remove air bubbles, place it at 16°C for 8-10 hours, and then store it at 4°C for future use.
Embodiment 3
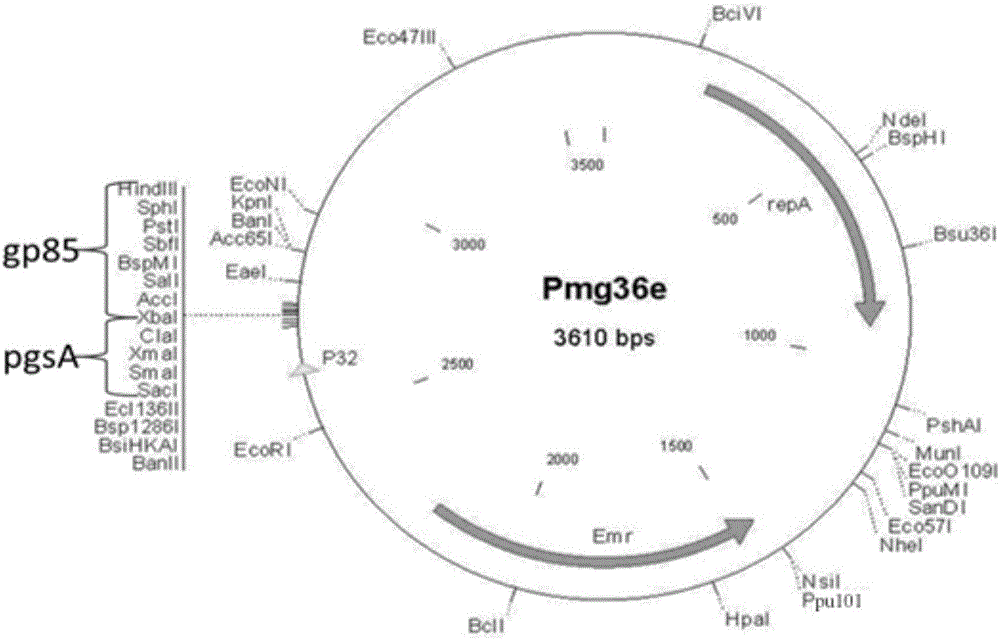
[0077] Example 3 Obtaining of recombinant plasmid pMG36e-pgsA-gp85
[0078] Recycling of expression vector pMG36e and gp85 target gene fragment by double enzyme digestion
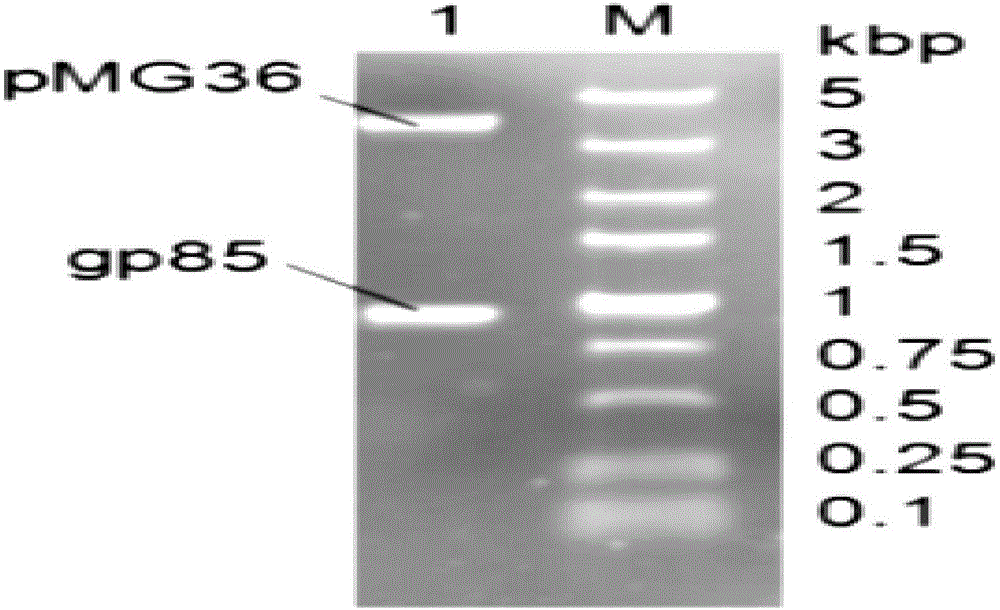
[0079] The expression vector pMG36e and the target gene gp85 fragment were digested with Hind Ⅲ and Xba Ⅰ respectively. Use 50 μl enzyme digestion system: 10×M Buffer, 6 μl; Xba Ⅰ and Hind Ⅲ 3 μl each; gp85 / pMG36e 15 μl; add ddH 2 O 23 μl. Mix the reactants according to the above system, and put them in a water bath at 37°C for 3h. The pMG36e vector fragment and the target fragment gp85 were purified and recovered with a gel recovery kit. After product recovery and purification, 1% agarose gel electrophoresis was used to observe the recovery situation.
[0080] Ligation transformation of target gene gp85 and expression vector pMG36e
[0081] a. Ligation: Ligate the enzyme-digested vector fragment recovered from the gel with the enzyme-digested target fragment. Use 20 μl ligation system: pMG36e 8 μl; g...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com