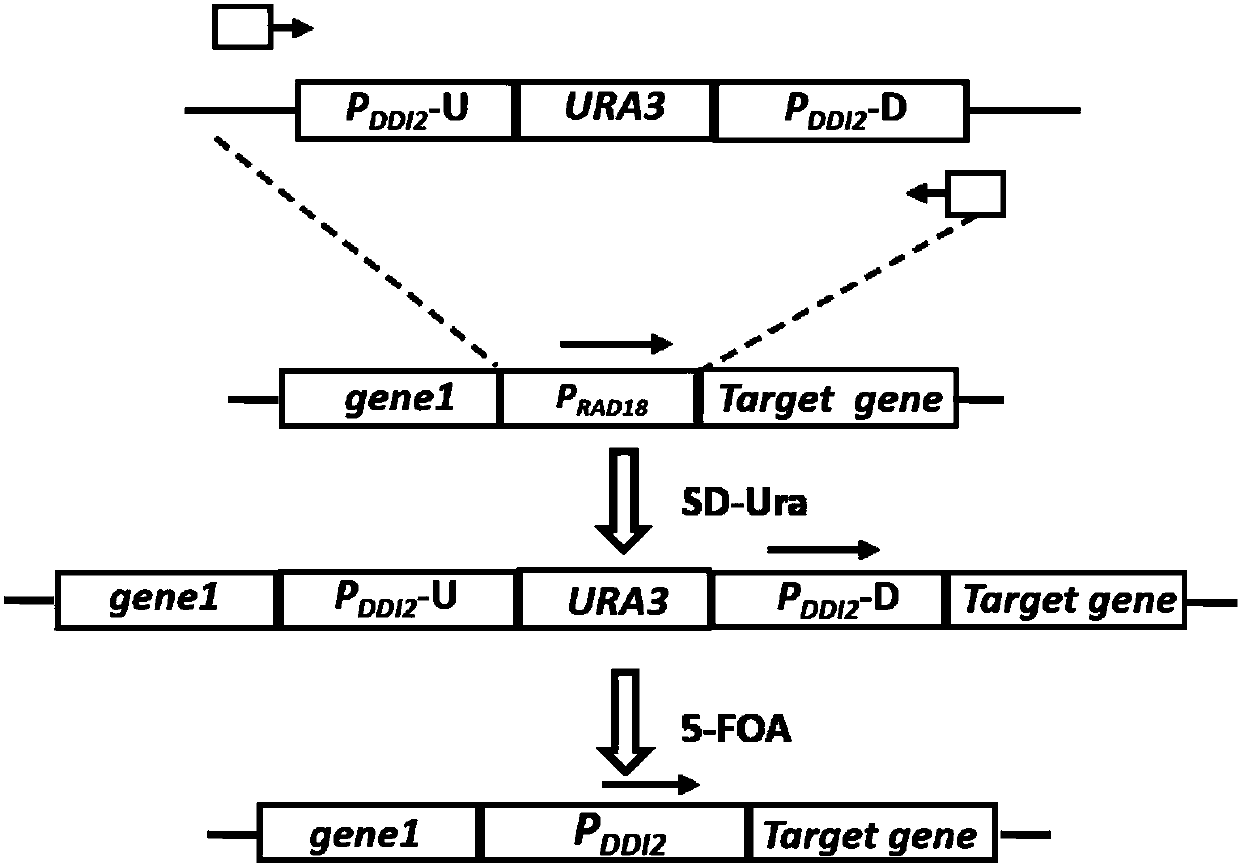
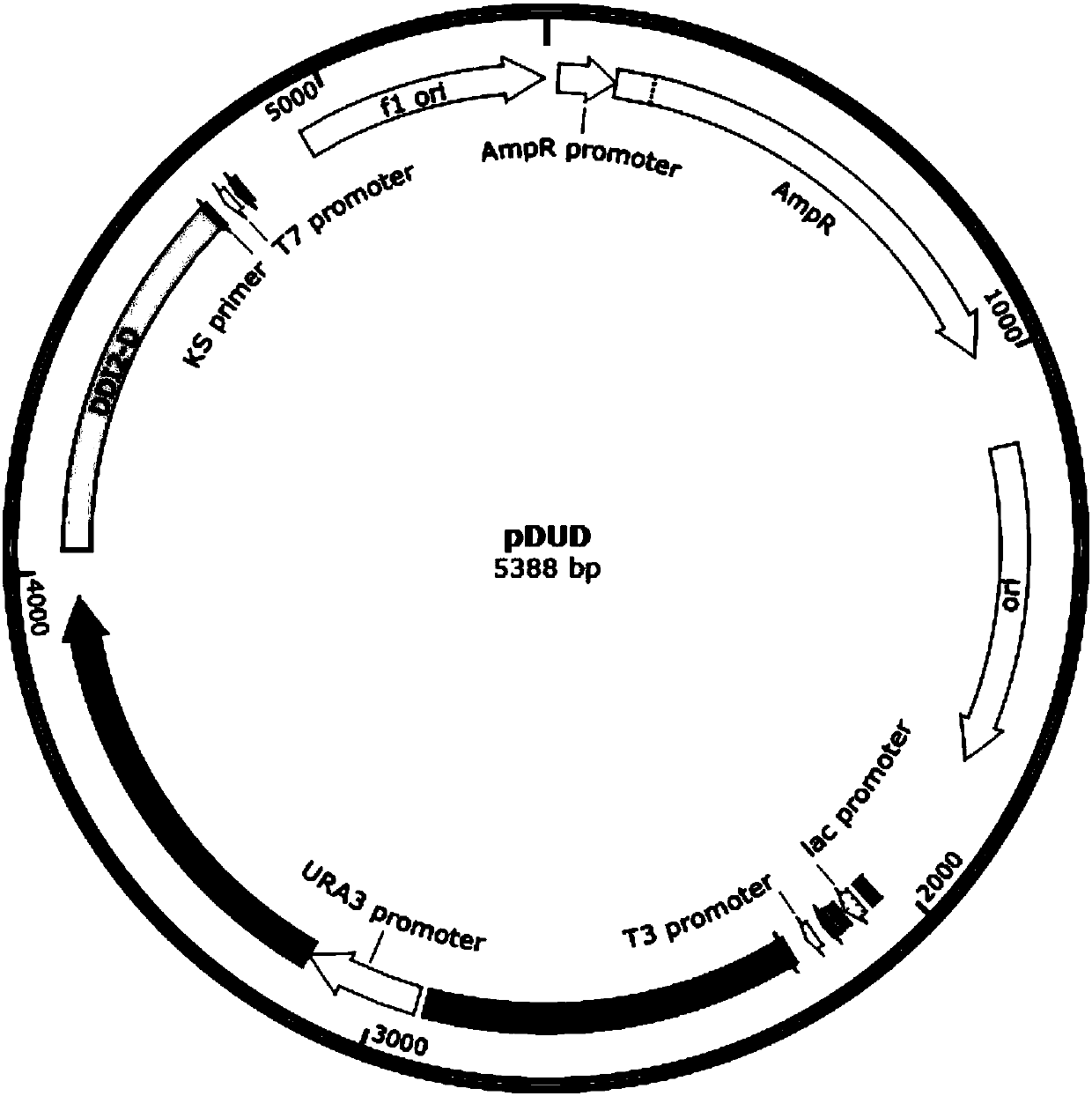
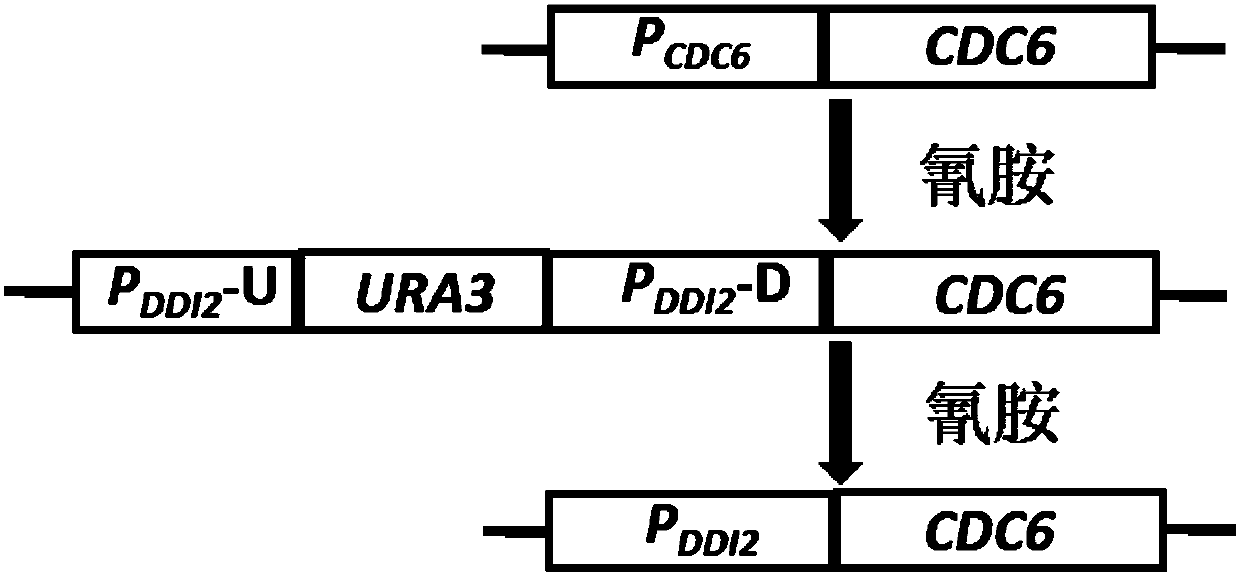
Method for replacing essential gene promoter in yeast genome
An essential gene and promoter technology is applied in the field of replacing essential gene promoters in yeast genome, which can solve the problems of difficult removal and ineffective regulation of essential genes, and achieves the effects of avoiding residues, facilitating the construction of strains, and realizing repeated use.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1DDI2
[0056] Example 1 DDI2 promoter seamlessly replaces yeast essential gene CDC6 promoter
[0057] 1. Construction of "DDI2-URA3-DDI2" repeat sequence plasmid
[0058] 1. Digest the plasmid pBlueScript-URA3 with restriction endonucleases BamHI and EcoRI, and recover the digested product of the plasmid;
[0059] 2. Amplify the promoter P with Phusion enzyme DDI2-U , recovering the PCR product;
[0060] 3. Ligate the vector backbone of step 1 and the recovered product of step 2 with Quick-Fusion enzyme to obtain recombinant plasmid pBlueScript-P DDI2-U -URA3, for sequencing verification.
[0061] 4. Then digest the plasmid pBlueScript-P with restriction enzymes HindIII and SalI DDI2-U -URA3, recovery of plasmid digestion products;
[0062] 5. Amplify the promoter P with Phusion enzyme DDI2-D ; Recover the PCR product;
[0063] 6. Ligate the vector backbone of step 4 and the recovered product of step 5 with Quick-Fusion enzyme to obtain recombinant plasmid pBlueScript-P DDI2-...
Embodiment 2
[0081] Example 2 Coated plate verification P DDI2 Replacement yeast essential gene CDC6 promoter strain
[0082] 1. Cultivate the strains obtained above in YPD-5mM cyanamide medium at 30°C overnight, and then transfer them to YPD / YPD-5mM medium the next day for 4 hours;
[0083] 2. Take the bacterial solution to measure the OD value. After calculation, take 107 bacterial solution, and after gradient dilution to 104 / 105, take 100 μL and 50 μL to smear the plate respectively, such as Figure 4 Shown, respectively 104, 1000, 500 bacteria;
[0084] 3. After coating on YPD / YPD-5mM plates, culture at 30°C for 2-4 days. Such as Figure 5 As shown, no plaques grew on the medium without cyanamide, indicating that P DDI2 The promoters of essential genes in yeast can be replaced by the above methods, and the expression of essential genes can be strictly regulated by inducers.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com