Corn salt-resistant main-effect QTL (Quantitative Trait Loci) as well as related genes, molecular markers and application thereof
A molecular marker, corn technology, applied in the field of genetic engineering to achieve the effect of accelerating the breeding process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Example 1 Cloning and Functional Analysis of Maize Salt Tolerance QTL Genes
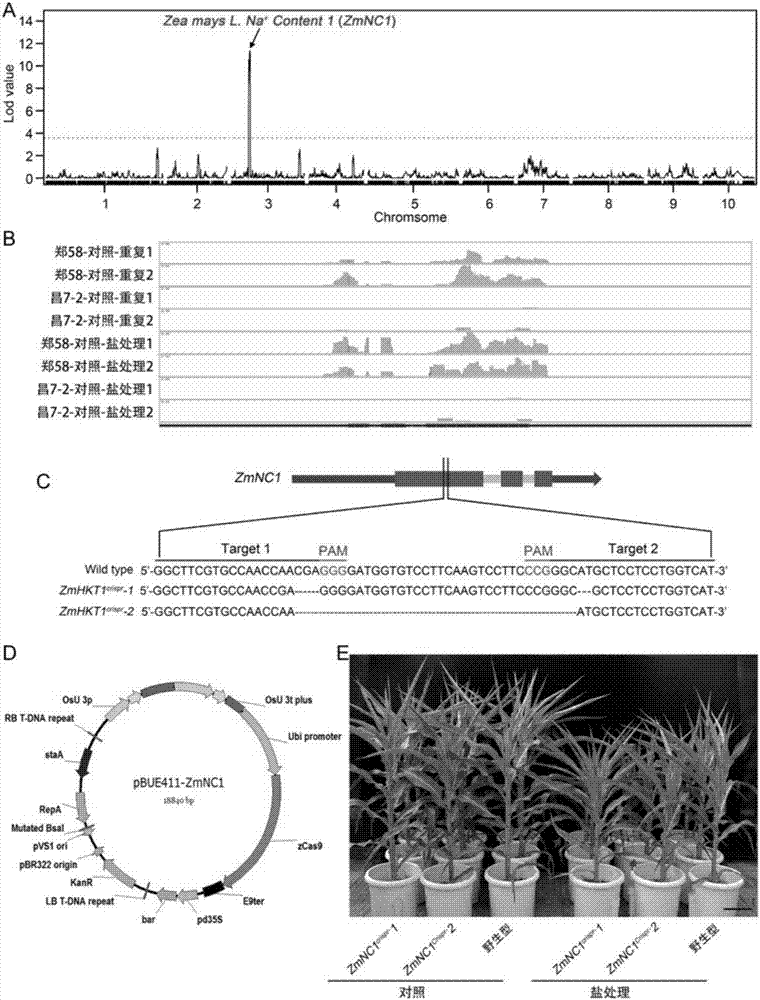
[0045] The salt resistance of the two parental inbred lines of Zhengdan 958, Zheng 58 and Chang 7-2, were significantly different, such as figure 1 shown. Using the Zheng 58 / Chang 7-2 RIL population as material, the present invention locates a major salt-resistance QTL region on chromosome 3 through QTL analysis based on a mixed linear model, such as figure 2 As shown in A, at the position of 55,028-55030kb on chromosome 3, the position is determined according to the maize B73 reference genome V2. On this basis, the transcriptomes of Zheng 58 and Chang 7-2 were compared between the control and salt stress by transcriptome sequencing, and a candidate gene was identified in this QTL candidate region, named ZmNC1, whose expression level was significantly lower in Chang 7-2 Yu Zheng 58, such as figure 2 As shown in B, the results showed that the expression level of this candidate gene in Chan...
Embodiment 2
[0052] Example 2 Obtaining a Large Inserted Fragment (ZmNC1-InDel) in the Exon of the Maize Salt Tolerance QTL Gene ZmNC1
[0053] The genomic DNA of Zheng 58 and Chang 7-2 was sequenced using the Illumina sequencing platform (sequencing was completed in Beijing Nuohezhiyuan Technology Co., Ltd.), and by comparing the whole genome sequencing data of Zheng 58 and Chang 7-2, it was found that In the salt-sensitive inbred line Chang 7-2, the second exon of the ZmNC1 gene has a large insertion, named ZmNC1-InDel, such as Figure 7 As shown in A. According to the whole genome sequencing data, the inventor designed two pairs of primers ZmNC1-F1 / ZmNC1-InDel-R1 and ZmNC1-InDel-F1 / ZmNC1-R1 (the specific positions of the primers on the gene are as follows: Figure 7 shown in A). The primer sequences are:
[0054] ZmNC1-F1: TACCTGCACACATCGATCGA;
[0055] ZmNC1-InDel-R1: CAACAGGAAATGGGCTGGAC;
[0056]ZmNC1-InDel-F1: CCAGAACACACCAGGAACCA;
[0057] ZmNC1-R1: TCAGGTTGATCGAGCGAGTT.
[...
Embodiment 3
[0059] Example 3 ZmNC1-InDel insertion or deletion in maize salt resistance gene ZmNC1 as a molecular marker
[0060] Because the ZmNC1-InDel insertion only exists in the salt-sensitive maize inbred line Chang 7-2, it causes a frameshift in the reading frame of the salt-resistant gene ZmNC1 and premature termination of translation, so the ZmNC1-InDel insertion can be used to determine whether an individual is salt-tolerant molecular markers.
[0061] Using primers ZmNC1-F1 (forward) and ZmNC1-R1 (reverse) designed based on the flanking sequence of ZmNC1-InDel insertion, and primer ZmNC1-InDel-R1 (reverse) designed based on the ZmNC1-InDel insertion sequence, to the above Three primers constitute primer pair I (ZmNC1-F1 / ZmNC1-R1) and primer pair II (ZmNC1-F1 / ZmNC1-InDel-R1). Using primer pair Ⅰ and Ⅱ as primers, the genome DNA of the salt-tolerant maize inbred line Zheng 58 and the salt-sensitive maize inbred line Chang 7-2 were respectively used as templates for PCR amplifica...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com