Screening, identification and application of SmAP2/ERF82 transcription factor for regulating tanshinone biosynthesis
A technology of ERF82, biosynthesis, applied in the field of plant molecular biology and genetic engineering
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
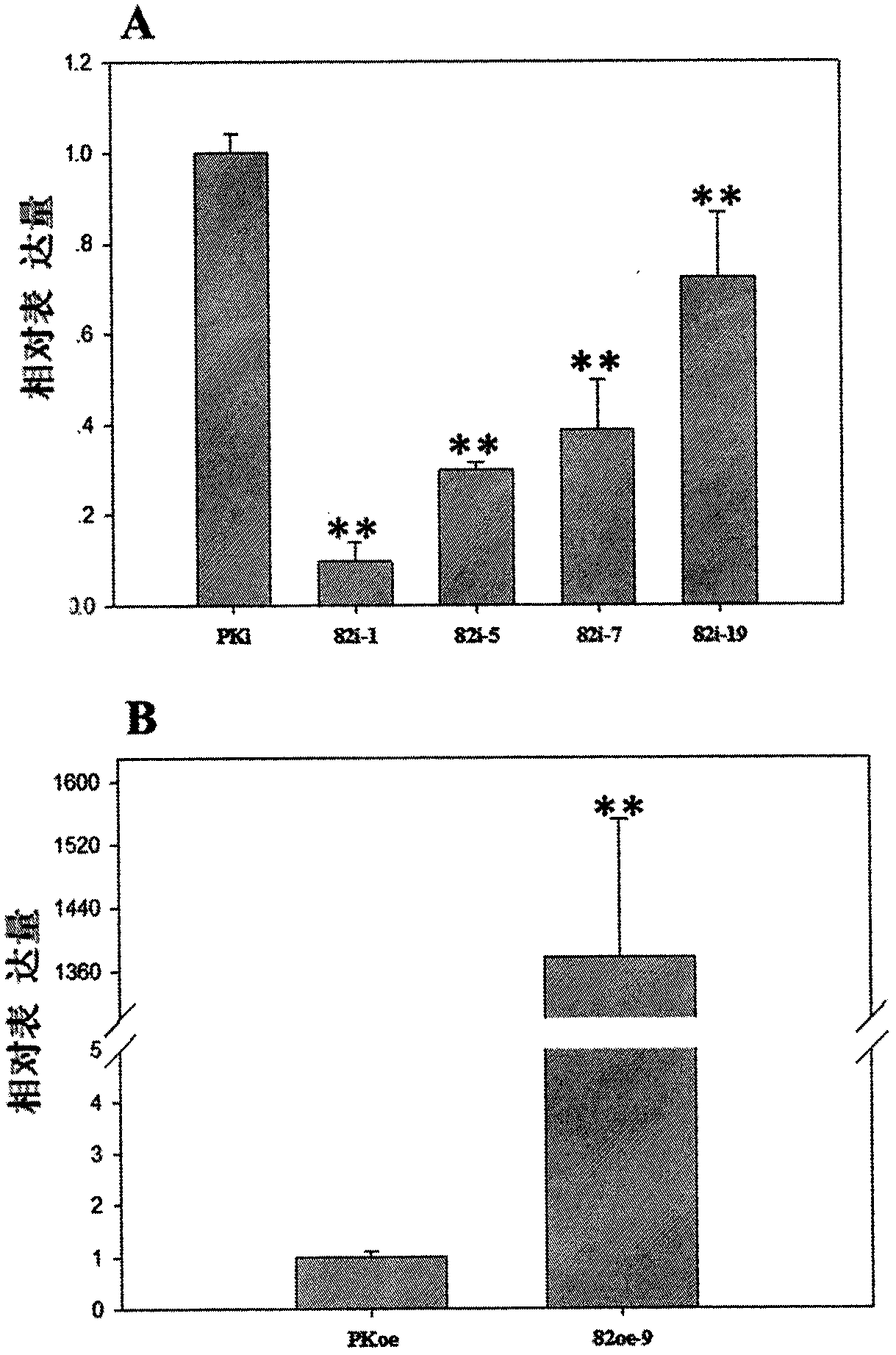
[0018] Example 1 Screening and identifying members of the AP2 / ERF gene family in the whole genome of Salvia miltiorrhiza
[0019] The genome of Salvia miltiorrhiza was searched and annotated using the hidden Markov model HMM: PF00847 in the Pfam database. A total of 170 AP2 / ERF transcription factor genes were predicted in the Danshen genome, named Sm001-Sm170, and the protein length ranged from 79aa to 595aa. Among them, Sm082 is named SmAP2 / ERF82 in the present invention.
Embodiment 2
[0020] Cloning of embodiment 2 Salvia miltiorrhiza SmAP2 / ERF82 gene
[0021] Design full-length primers according to the open reading frame of the SmAP2 / ERF82 sequence, use the cDNA of Salvia miltiorrhiza as a template, and PCR amplify to obtain the nucleotide sequence of the SmAP2 / ERF82 gene, which is 582 bp in length, such as SEQ ID No.1. The amino acid sequence of SmAP2 / ERF82 was deduced after translation of the nucleotide sequence, including 193 amino acid residues, such as SEQ ID No.2.
Embodiment 3
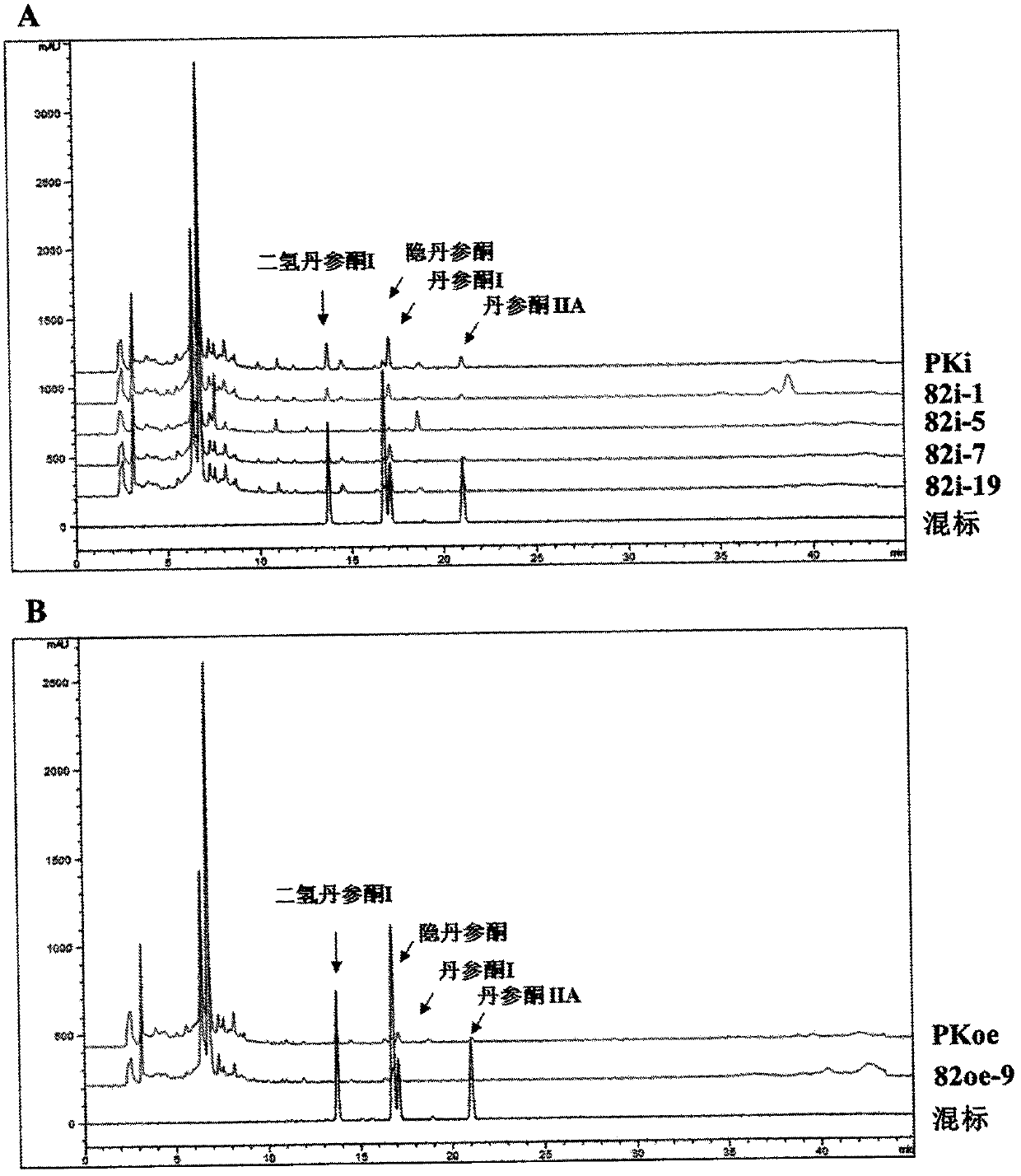
[0022] Example 3 Functional verification of Danshen SmAP2 / ERF82 gene
[0023] 1) RNAi primer design and PCR amplification. Select a 185bp specific fragment in the SmAP2 / ERF82 gene as the RNAi target region (located at 96-280bp of the gene), design primers at both ends of the target region, and add attB sequence at the 5' end of the primer according to the principle of Gateway, where The F primer adds the attB1 sequence: GGGGACAAGTTTGTACAAAAAAAGCAGGCT, and the R primer adds the attB2 sequence: GGGGACCACTTTGTACAAGAAAGCTGGGT. The primer sequences are as follows:
[0024] F- GGGGACAAGTTTGTACAAAAAAGCAGGCT GATCGCCGCCGTTGTCGGC
[0025] R- GGGGACCACTTTGTACAAGAAAGCTGGGT TGTCGAATGTGCCGAGCCA
[0026] 2) Overexpression primer design and PCR amplification. The attB sequence was added to the 5' end of the full-length primer of the SmAP2 / ERF82 gene. The primer sequences are as follows:
[0027] F- GGGGACAAGTTTGTACAAAAAAGCAGGCT ATGTTGAGAAACGACTCTTT
[0028] R- GGGGACCACTTTGTACA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com