Two pairs of specific primers for detecting acid-resistant lactobacillus content during Baijiu fermentation and application thereof
A fermentation process, Lactobacillus technology, applied in the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc. Conducive to quality control, advanced design methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Extraction of metagenomics from pit mud or fermented grains
[0026] Use the Shanghai Sangon B518233-0100 kit to extract the metagenome, weigh 100-300 mg of cellar mud (if it is wine grains, you need to crush it in advance, and weigh 100-300 mg after crushing), add 400ul Buffer SCL preheated at 65°C, Shake well and place in a 65°C water bath for 5 minutes. Centrifuge at room temperature at 12000rpm for 3min, and pipette the supernatant into a clean 1.5ml centrifuge tube. Add an equal volume of Buffer SP, invert to mix, and ice-bath for 10 minutes. After adding Buffer SP and mixing, the solution becomes white or light yellow turbid. Centrifuge at room temperature at 12000rpm for 3min, and pipette the supernatant into a clean 1.5ml centrifuge tube. Add 200ul of chloroform, mix well, centrifuge at 12000rpm for 5min, draw the upper layer of the aqueous phase into a clean 1.5ml centrifuge tube. Add 1.5 times the volume of Buffer SB, mix well, add all of it to the adsorpti...
Embodiment 2
[0028]Primer design and amplification of target DNA
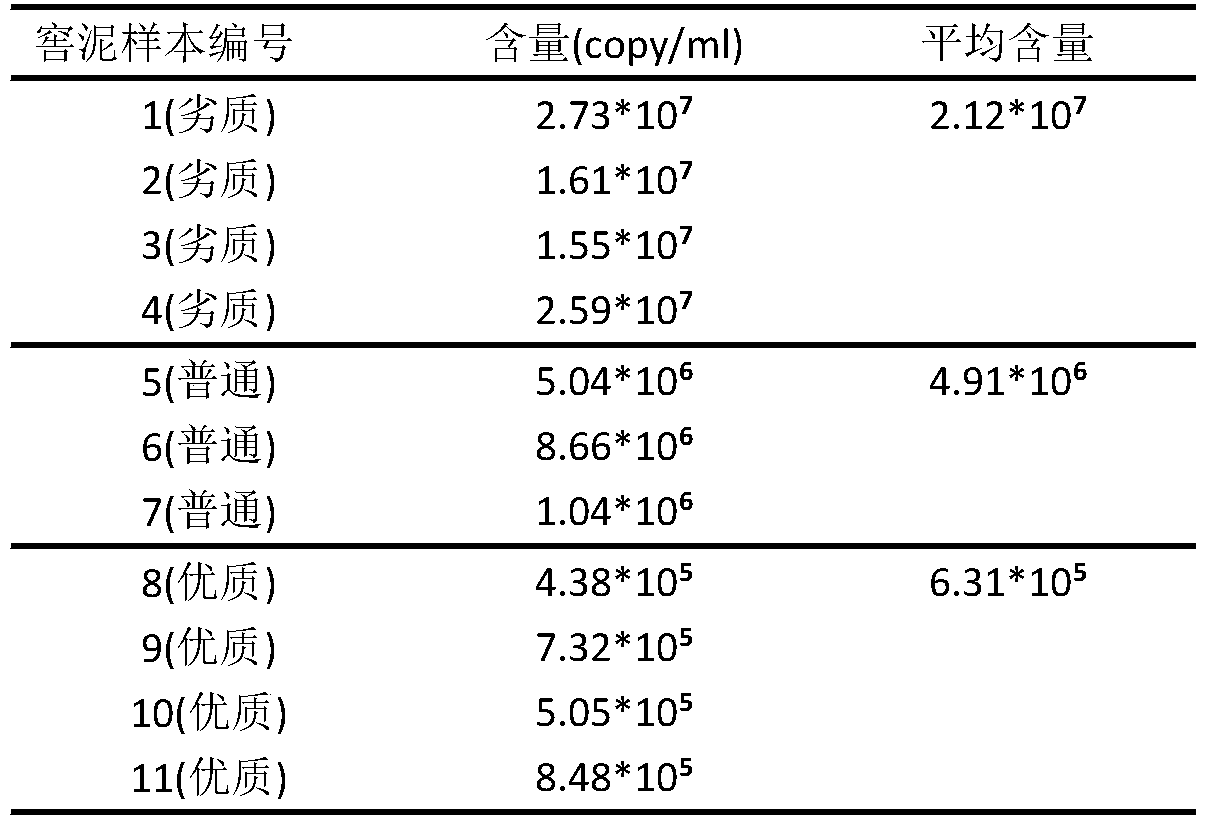
[0029] Use Primer-Blast to design species-specific primers for the whole genome of Lactobacillus acetotolerans. The purpose of designing specific primers for a certain species is generally based on two reasons. One is to identify the species and detect whether the species is contained in a certain biological community. Another aim was to determine the abundance of the species in the overall microbial community.
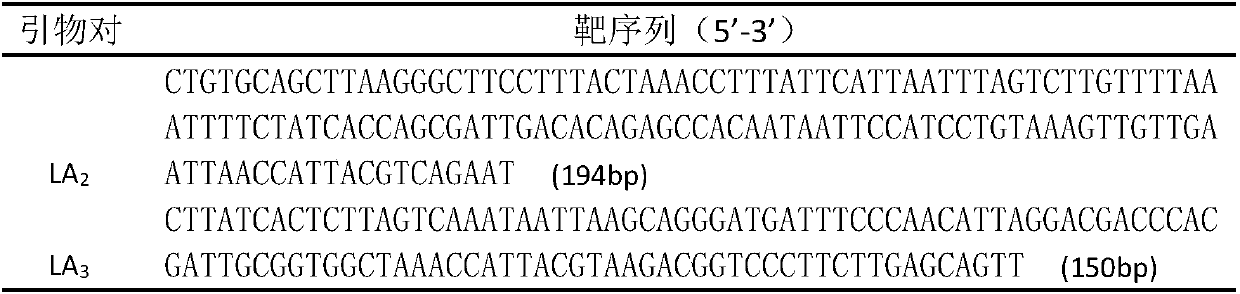
[0030] Table 1 Species-specific primers for Lactobacillus acetotolerans
[0031]
[0032] Table 2 Amplified sequences of two pairs of primers
[0033]
[0034] PCR reaction system: NPK02buffer (2×) 6ul, species-specific primer LA2F LA2R (or LA3F LA3R) 1.5ul, template 0.15ul (use fermented grains metagenomic mixed sample as template), Taq enzyme 0.25ul, ddH2O 5.1ul. 94°C-4min, (94°C-30s, 57°C-40s, 72°C-40s)×35 cycles, 72°C-2min 37 cycles. The purified PCR products were used for TA cloning and blue-white scr...
Embodiment 3
[0036] Validation of primer specificity by TA cloning
[0037] PMD19-T vector 0.3ul, LA2 (or LA3) PCR purified product 0.5ul, sterilized ddH2O 4.2ulSolution I 5ul, mix well, add 30ul Escherichia coli competent cells DH5α ice bath for 40 minutes, then shake at 37°C Incubate and activate for 1 h. Mix X-gal 40ul, IPTG 40ul, 160ul dH2O, spread on LB ampicillin (100ug / ml) solid medium (LBA), wait for it to dry, continue to spread 200ul of activated bacteria solution on X-gal+IPTG Dried LBA solid medium. Incubate overnight at a constant temperature of 37°C. Randomly pick 15 single colonies and send them to the sequencing company for full-length sequencing of the inserted sequence. The sequencing results were subjected to BLAST analysis and RDP analysis (http: / / rdp.cme.msu.edu / ) to conduct qualitative and quantitative analysis on whether the amplified product belonged to the target Lactobacillus. All 15 cloned sequences are target sequences to pass the specificity test.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com