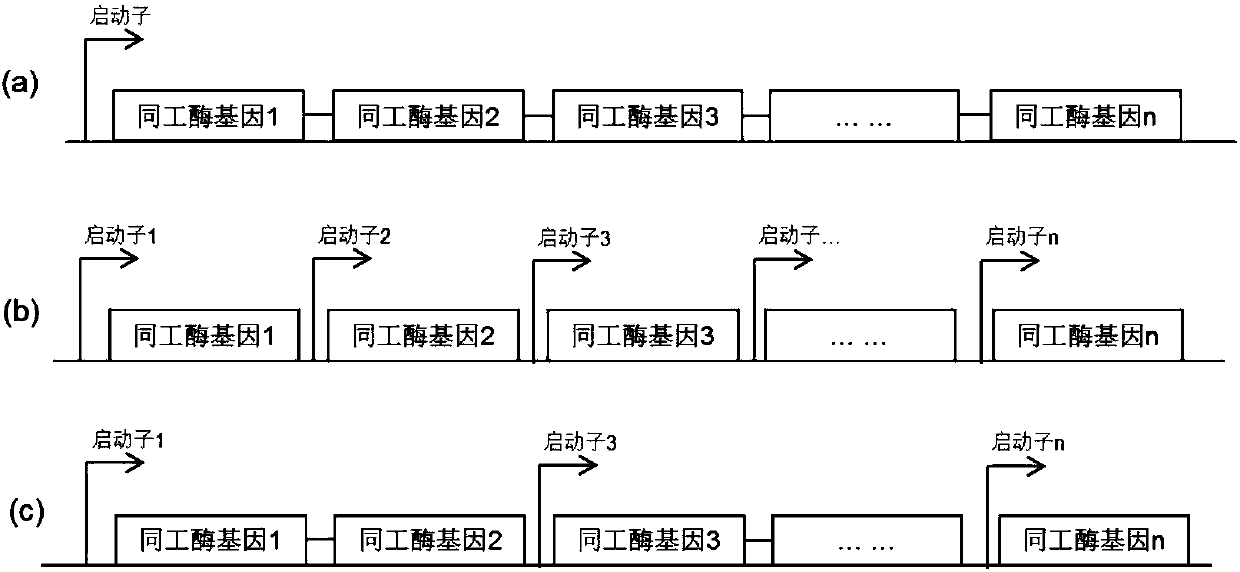
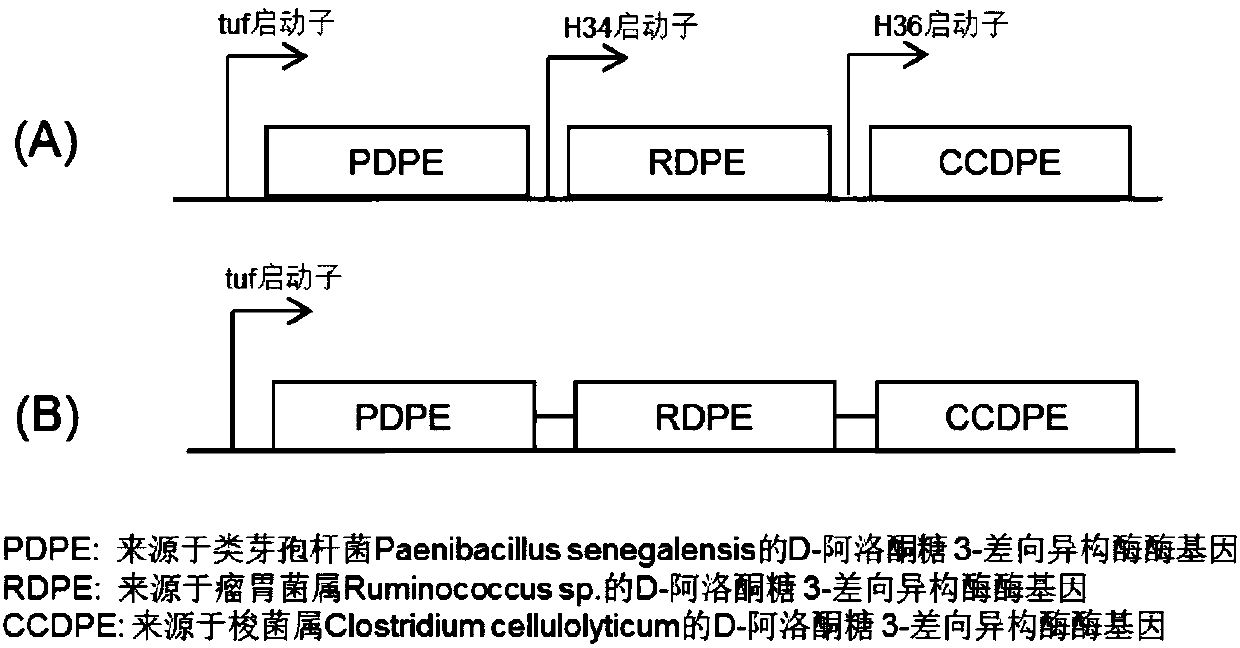
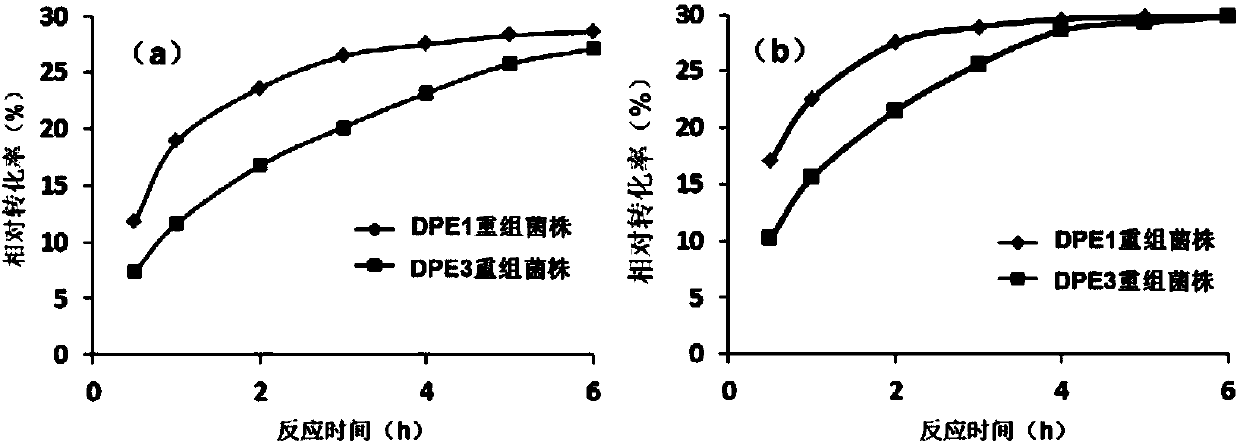
Method for efficiently preparing D-psicose 3-epimerase and use of D-psicose 3-epimerase
A technology for epimerase and psicose, which is applied in the efficient preparation of D-psicose 3-epimerase and the application field in the preparation of psicose, and can solve the problems of low reaction efficiency and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0073] Embodiment 1, construction Corynebacterium glutamicum recombinant strain expresses combined DPE operon with the form of episomal plasmid
[0074] 1. Construction of recombinant strain DPE1 of Corynebacterium glutamicum
[0075] First, construct the recombinant expression vector pEC-RPCDPE1 and design primers 1 and 2 according to the tuf promoter sequence (SEQ ID NO: 4) derived from Corynebacterium glutamicum, with the DPE gene sequence (SEQ ID NO: 1) derived from Paenibacillus senegalensis ) designed primers 3 and 4, designed primers 5 and 6 with the DPE gene sequence (SEQ ID NO: 2) derived from Ruminococcus sp., and designed primers 7 and 6 with the DPE gene sequence (SEQ ID NO: 3) derived from Clostridium cellulolyticum 8, wherein primers 2 and 3, primers 4 and 5, and primers 6 and 7 respectively contain a homologous region of about 40 bp for fusion between PCR fragments, primer 5 contains the H34 promoter sequence, and primer 7 contains the H36 promoter subsequence,...
Embodiment 2
[0109] The expression of the combined DPE operon of embodiment 2, plasmid form in Corynebacterium glutamicum
[0110] 1. Cultivation of recombinant strains of Corynebacterium glutamicum
[0111] Select 100mL BHI medium (brain heart extract powder 37g / L, kanamycin 25ng / mL), and cultivate the recombinant strains DPE1 and DPE2 of Corynebacterium glutamicum at 30°C and 200rmp for 12-24h, 4°C, Centrifuge at 8000rmp for 15min to collect the bacteria, and use ddH 2 O Concentrate the bacterial solution to 5 mL, and prepare a crude enzyme solution by ultrasonic crushing.
[0112] 2. Determination of enzyme activity
[0113] In order to measure the enzyme activity of crude enzyme, set up the following reaction system (500ul): D-fructose: 5%, crude enzyme: 0.1mg, MnCl 2 : 1mM; react for 10min, add NaOH to terminate the reaction, and perform liquid chromatography detection.
[0114] The crude enzyme activity of combined DPE and single DPE was analyzed respectively by using the above s...
Embodiment 3
[0115] Example 3, construction of recombinant strains of Corynebacterium glutamicum to express combined DPE operons in the form of chromosomal integration
[0116] 1. Construction of the recombinant strain DPE3 of Corynebacterium glutamicum
[0117] First, the integration site vector pK18-ldhA was constructed to integrate the DPE combined operon into the lactate dehydrogenase gene (ldhA) site of the chromosome, according to the upstream and downstream sequence information of the lactate dehydrogenase gene (ldhA) in Corynebacterium glutamicum , design primer 9, primer 10, primer 11 and primer 12, wherein primer 9 and primer 10 are used to amplify the upstream gene fragment of ldhA, primer 11 and primer 12 are used to amplify the downstream gene fragment of ldhA, primer 10 and primer 11 There is a homologous region of about 40 bp, which is used for the fusion of upstream and downstream fragments. Primer 9 and primer 12 contain EcoRI and HindIII restriction sites respectively. T...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com