A Nucleic Acid Mass Spectrometry Parentage Testing Method Based on Informational SNP Set and Its Primers
A technology of paternity identification and nucleic acid mass spectrometry, which is applied in the fields of 30 information SNP marker combinations, primer sequences and paternity identification, can solve the problems of interpretation of results, affect the interpretation of paternity identification results, and difficult DNA typing, etc., and achieve high accuracy high effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
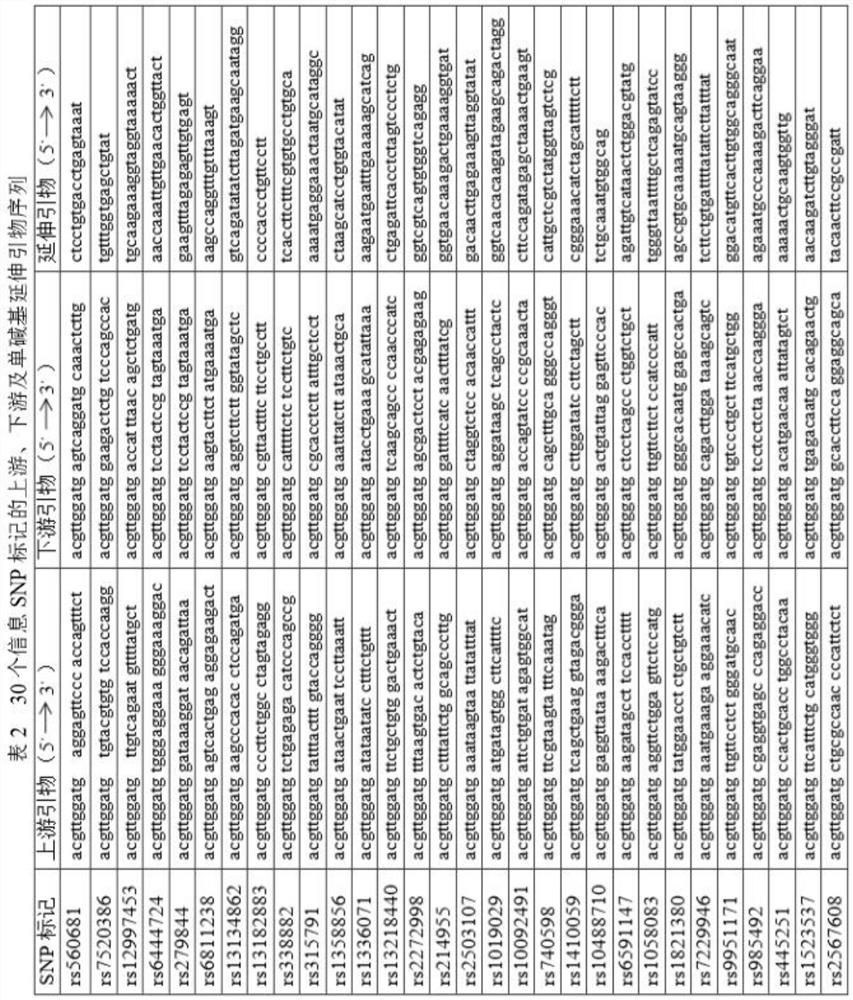
[0016] Example 1: Selection of 30 informative SNP markers
[0017] The selection of the 30 information SNP markers used in the paternity testing system in the present invention is based on the data analysis of 2100 individuals mainly from African, European, American and Asian populations, and has gone through three processes, specifically as follows:
[0018] 1) The number of SNPs with heterozygous rate>0.45 and Fst<0.01 was initially screened out from the population data SNPs is 2723;
[0019] 2) On the basis of selecting high heterozygosity rate and low Fst index, not selecting SNPs with a position distance of less than 1Mb, and not selecting sites on the X or Y chromosomes, 195 SNPs for primary screening were obtained;
[0020] 3) SNPs with heterozygosity rate>0.4, population genetic index Fst<0.06 and mutation frequency<0.01% were screened among 195 SNPs initially screened, and 30 optimal SNPs as shown in Table 1 were selected.
Embodiment 2
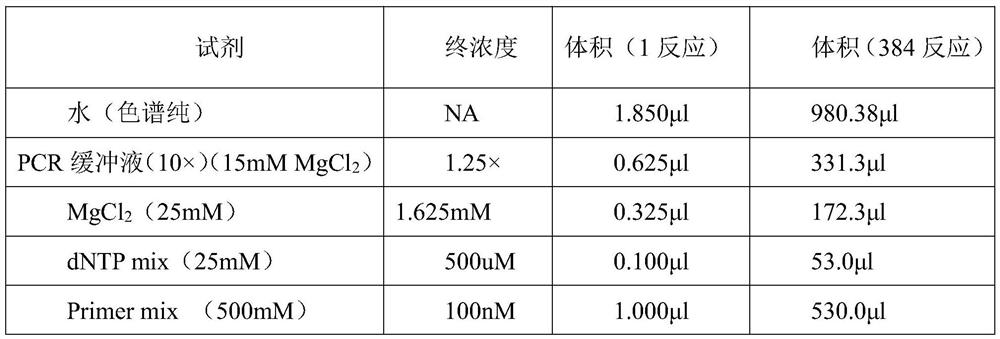
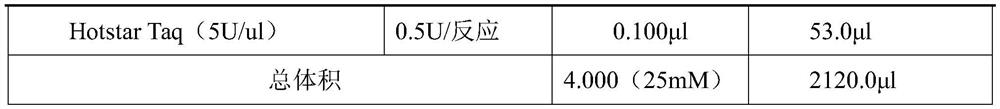
[0021] Example 2: Parentage identification based on nucleic acid mass spectrometry detection information SNP set
[0022] 1. Genome Extraction
[0023] In this embodiment, the magnetic bead method is used to extract genomic DNA. The sample is saliva, and 2ml of saliva is collected and stored in a saliva stabilizer. The extraction steps of saliva genomic DNA are as follows:
[0024] 1) Add 450 μl Lysis Buffer V, 450 μl sample (mixture of saliva and saliva preservation solution), 220 μl isopropanol, 20 μl proteinase K and 10 μl nucleic acid sedimentation aid to a 1.5ml centrifuge tube, vortex and mix well, and place the centrifuge tube Water bath at 56°C for 10 minutes, and invert 6-8 times during the period.
[0025] 2) Take out the centrifuge tube, add 20 μl of magnetic beads and vortex slightly for mixing, then place in a water bath at 56°C for 20 minutes, during which time vortex every 2 minutes.
[0026] 3) Take out the centrifuge tube and place it on the magnetic stand f...
Embodiment 3
[0071] Embodiment 3: Application in real groups
[0072] In order to verify the feasibility and accuracy of the entire set of information SNP sets for paternity testing in actual families, 20 groups of samples were collected in this example, of which 10 groups were combinations with a clear father-son relationship, and the remaining 10 groups were unrelated unknown males with any A child's pairing combination. These 20 groups of samples are numbered KY-1~KY-20 respectively. The data obtained in Example 2 were used to infer the parent-child relationship, and the results are shown in Table 10.
[0073] Table 10 Application of 30 informative SNP sets in practical populations
[0074] combination number Cumulative Parentage Index (CPI) Relative Chance of Paternity (RCP) actual parental relationship KY-1 856324110 1 biological KY-2 625341255.97 1 biological KY-3 965423826.91 1 biological KY-4 0 0 Not biological KY-5 0 0 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com