Single nucleotide polymorphism based primer set for hybrid identification of true and false peanuts and identification method thereof
A single nucleotide polymorphism and primer set technology, applied in biochemical equipment and methods, microbial measurement/testing, recombinant DNA technology, etc., can solve inaccurate detection results, low resolution of electrophoresis detection, and accurate identification results Reduce the speed and other problems, to save time and improve efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0042] Below in conjunction with embodiment the present invention is further elaborated.
[0043] 1. Selection of materials: 21 peanuts were selected, and the genomic DNA of the peanut samples was extracted using the BioTeke Magnetic Bead Method Plant Genomic DNA Extraction Kit (Cat#AU31111).
[0044] 2. Using peanut genomic DNA as a template, a round of PCR amplification was performed with the primer mixture to obtain the target region;
[0045] The one-round PCR amplification system: primer mixture 10 μl; DNA dosage 100 ng; 3*M enzyme 15 μl; add water to make up 45 μl.
[0046] The one-round PCR amplification system: 95°C for 3min; (95°C for 30s, 60°C for 4min, 72°C for 30s), 17 cycles; 72°C for 4min.
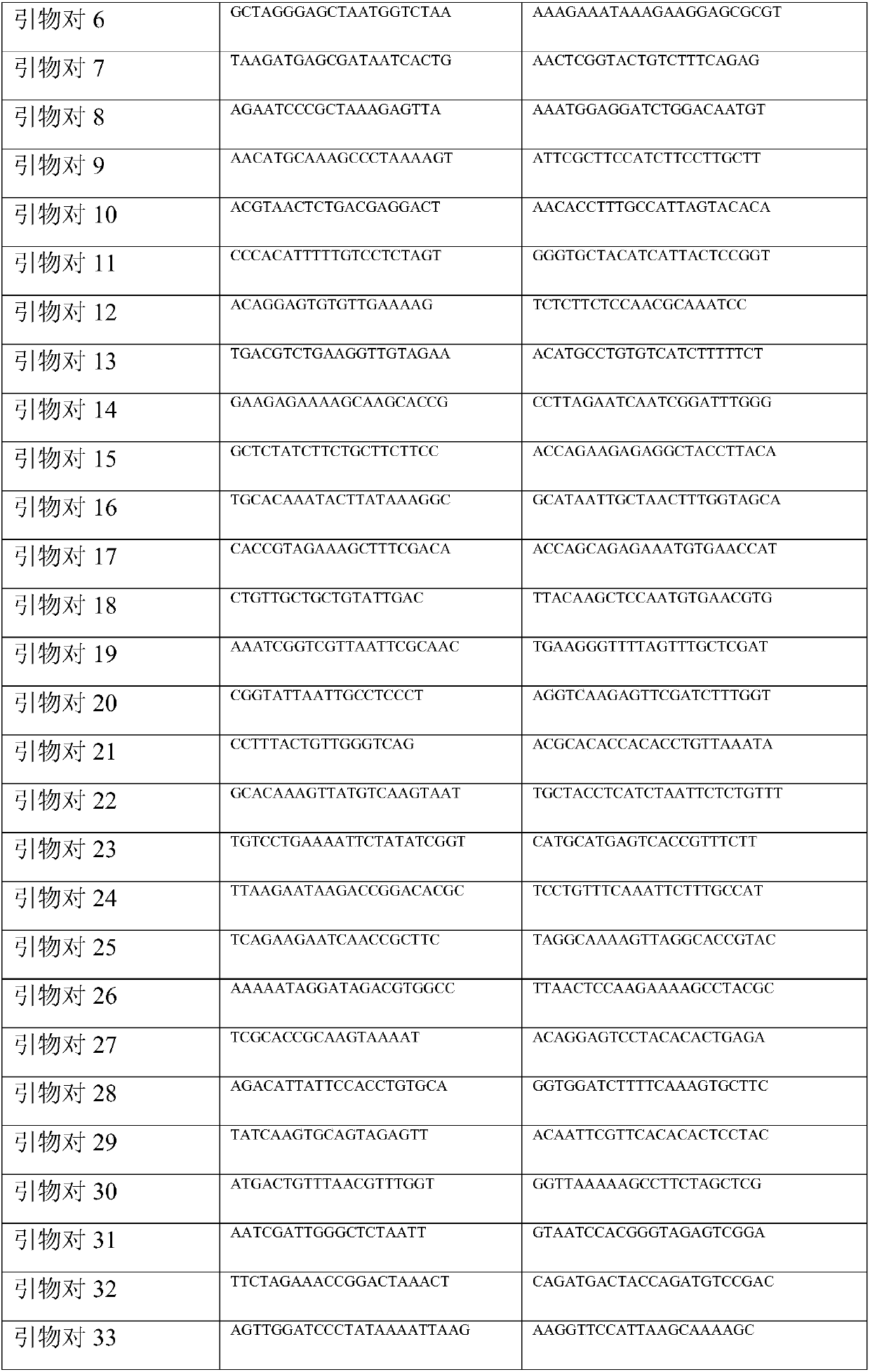
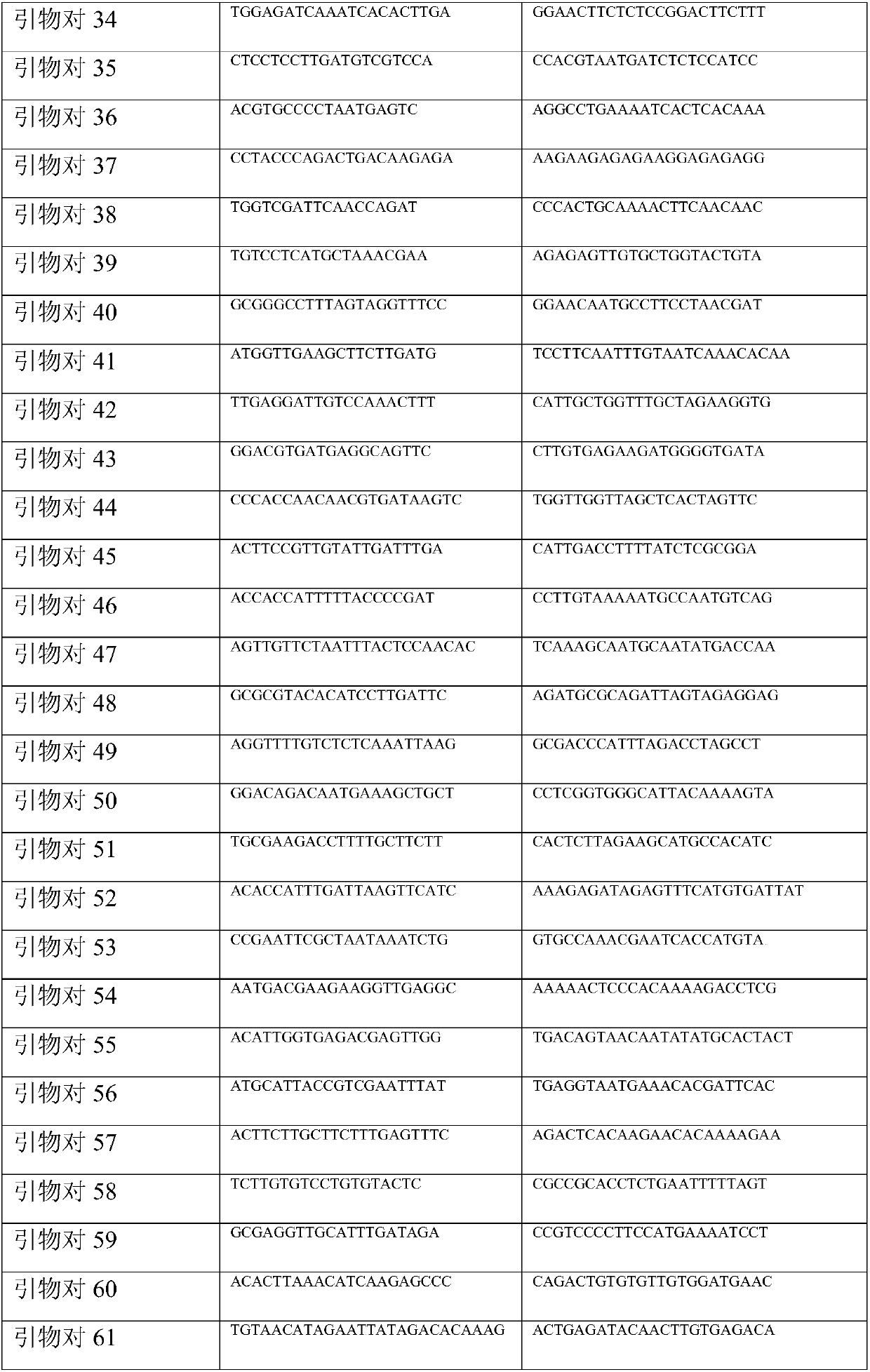
[0047]Primer mixture: there are a total of 136 primers in the present invention, as shown in Table 1. Take 10 μl of each primer and dilute to 10 ml. The concentration of each primer is 0.1 μM.
[0048] Table 1 Primer Set
[0049]
[0050]
[0051]
[0052]
[...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com