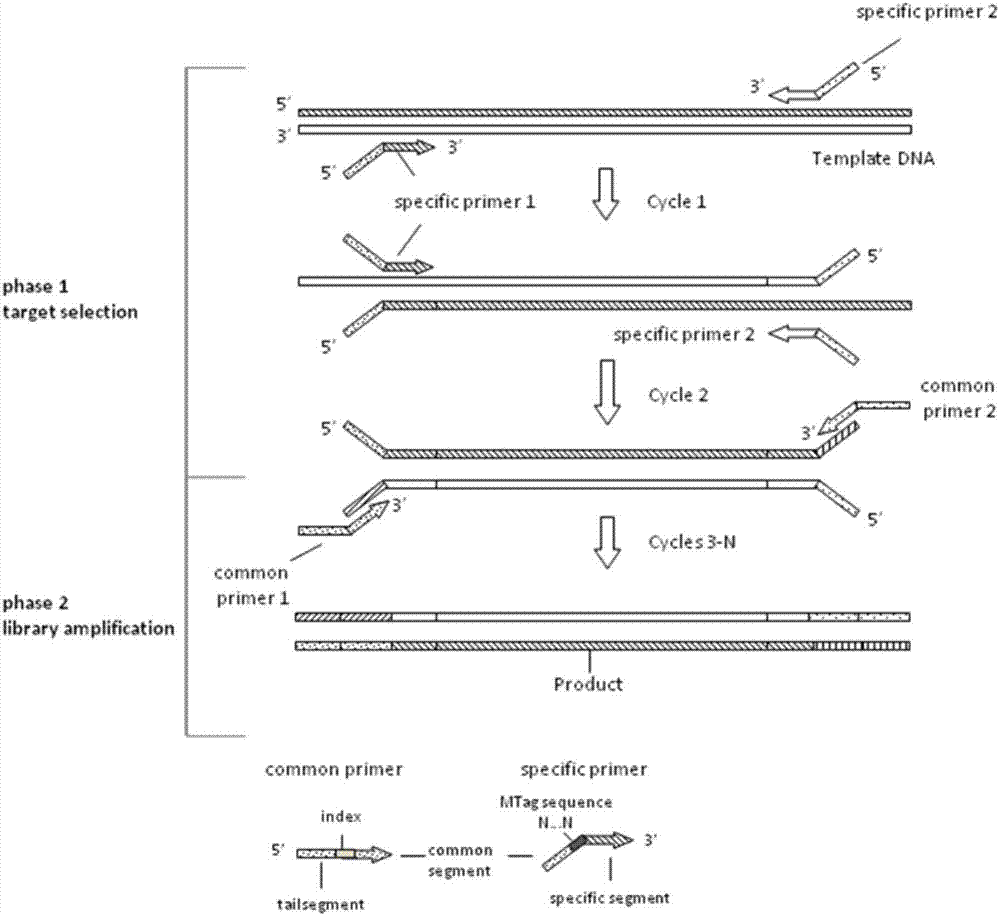
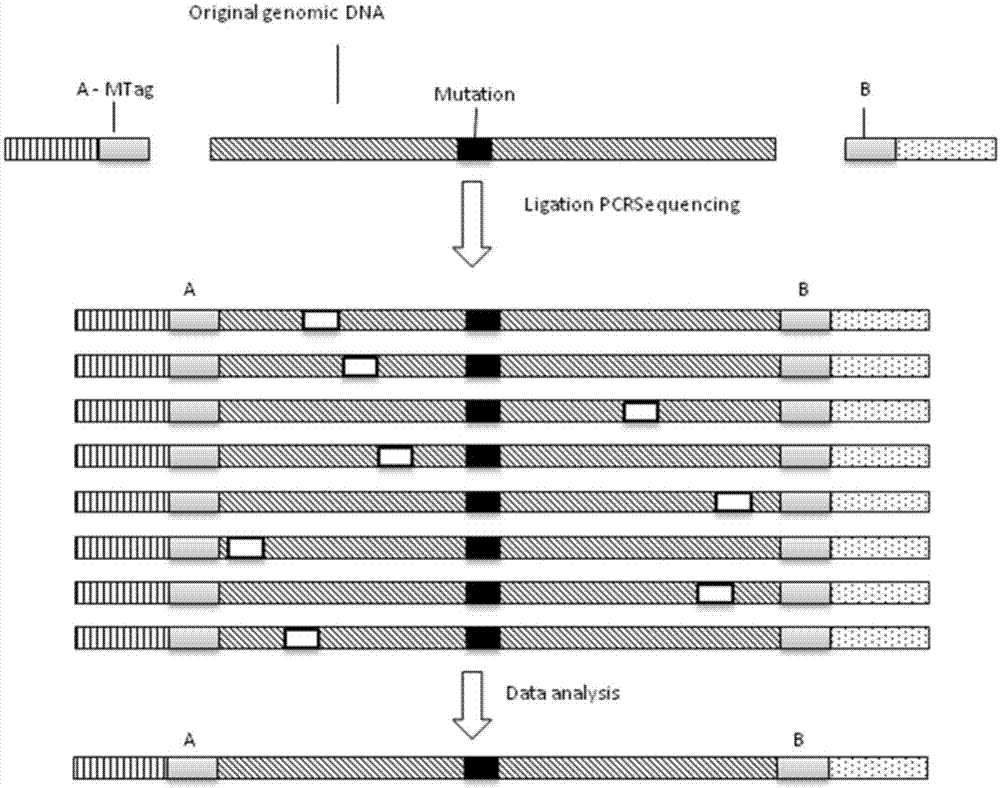
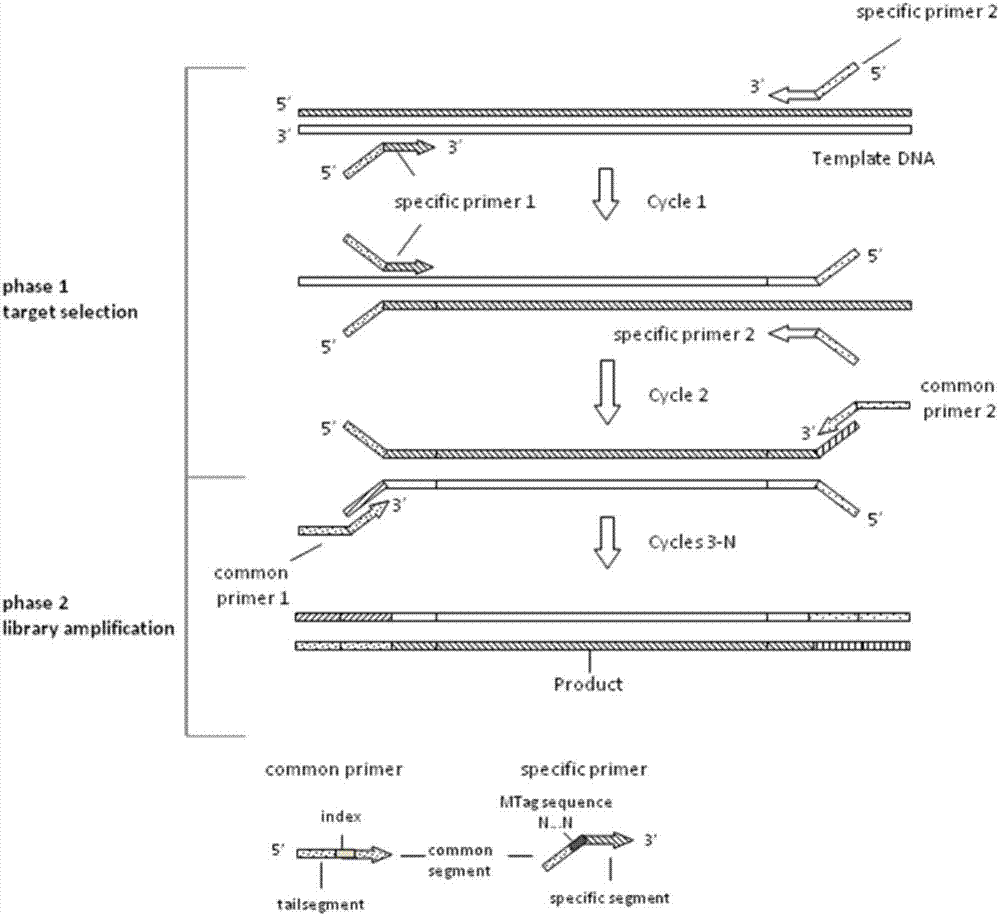
Kit for detecting colorectal cancer susceptibility gene mutation and detection method thereof
A technology of colorectal cancer and susceptibility genes, applied in biochemical equipment and methods, microbiological determination/testing, DNA/RNA fragments, etc., can solve specific sensitivity, lack of specificity, low detection sensitivity, etc. problems, to achieve a wide range of clinical applications, improve primer specificity, and improve detection sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] The reliability of the kit and method was verified by detecting the standard of known mutation type and frequency.
[0034] A. FFPE DNA extraction
[0035] Use the extraction kit QIAamp DNA FFPE Tissue Kit (Qiagen, Cat. No. 56404) to extract FFPE DNA from paraffin coils (Horizon, Cat. No. HD200), and the operation is as described in the kit instruction manual. The resulting product is used Fluorescence Quantitative Instrument and Corresponding The quantitative reagent (Invitrogen) was used for quantification, and the calculated concentration of the extracted FFPEDNA was 20 ng / μL.
[0036] B. multiplex PCR method to amplify the target region of the genome of the sample to be tested.
[0037] 1). Prepare a sterile, nuclease-free 200 μL PCR tube and place it on ice; prepare the PCR reaction system as shown in the table below, and be careful to avoid cross-contamination.
[0038]
[0039] The basic information of the primers is as follows, N means A, C, G or T:
...
Embodiment 2
[0064] The reliability of the kit is verified by comparing the mutation type and frequency of the target genomic DNA in the patient's plasma with the results of digital PCR (ddPCR).
[0065] A. Extraction of target genomic DNA from patient plasma
[0066] Using the extraction kit QIAamp Circulating Nucleic Acid Kit (Qiagen, Cat. No. 55114), the target genomic DNA was extracted from the plasma of 5 patients, and the operation was as described in the instruction manual of the extraction kit. The resulting product is used Fluorescence Quantitative Instrument and Corresponding The quantitative reagent (Invitrogen) was used for quantification, and the calculated concentration of the extracted DNA was 15.0 ng / μL.
[0067] B. multiplex PCR method to amplify the target region of the genome of the sample to be tested.
[0068] 1). Prepare a sterile, nuclease-free 200 μL PCR tube and place it on ice; prepare the PCR reaction system as shown in the table below, and be careful to avo...
Embodiment 3
[0093] The reliability of the kit was verified by comparing the mutation type and frequency of DNA in FFPE samples of patients with digital PCR (ddPCR) results.
[0094] A. DNA Extraction from Patient FFPE Samples
[0095] The FFPE tissue samples of five patients were extracted using the extraction kit QIAamp DNA FFPE Tissue Kit (Qiagen, Cat. No. 56404), and the operation was as described in the instruction manual of the extraction kit. The resulting product is used Fluorescence Quantitative Instrument and Corresponding The quantitative reagent (Invitrogen) was used for quantification, and the calculated concentration of the extracted DNA was 24.0 ng / μL.
[0096] B. multiplex PCR method to amplify the target region of the genome of the sample to be tested.
[0097] 1). Prepare a sterile, nuclease-free 200 μL PCR tube and place it on ice; prepare the PCR reaction system as shown in the table below, and be careful to avoid cross-contamination.
[0098]
[0099]
[01...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com