Hyper-blocking fluorescence quantitative PCR method with high sensitivity for detecting rare mutation
A fluorescence quantitative and pass-through technology, applied in the field of clinical diagnosis and molecular biology, can solve the problems of high operator requirements, complicated operation process, and insufficient detection sensitivity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
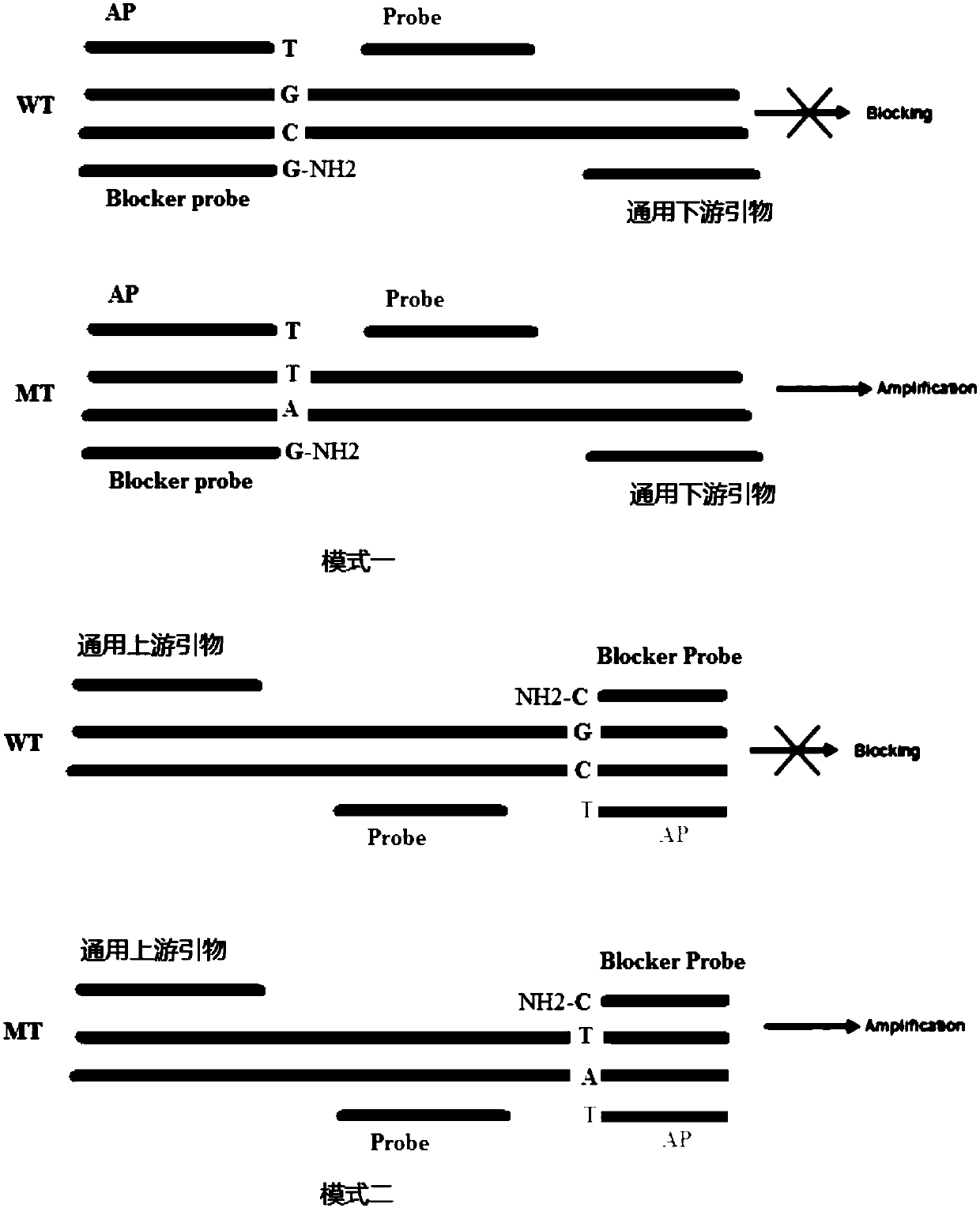
[0083] Embodiment 1, the design of ultra-retardation fluorescent quantitative PCR primer probe
[0084] The primer-probe combination used to detect rare gene mutation sites consists of an upstream universal primer F, a downstream universal primer R, a general detection probe P, a specific amplification primer AP, and a locked nucleic acid blocking probe BL.
[0085] The upstream universal primer F, the downstream universal primer R and the general detection probe P sequences are all derived from the wild-type and mutant homology regions of the detection gene; the upstream universal primer F and the downstream universal primer R are 13-25nt in length , the TM value is 56-66°C; the length of the pass-through probe P is 12-30nt, and the TM value is 66-72°C; the pass-through probe P is modified by LNA or MGB; the LNA modification The number is 1-5, and the 4 base monomers of ATCG can be modified with LNA, and the 5' and 3' ends are respectively labeled with a fluorescent group and...
Embodiment 2
[0090] Embodiment 2, establishment of ultra-retardation fluorescent quantitative PCR detection method
[0091] 1. Extract the total RNA of the sample to be tested and reverse transcribe it into cDNA.
[0092] 2. Using the cDNA obtained in step 1 as a template, carry out a control PCR reaction and a super-retardation PCR reaction;
[0093] Control PCR reaction system (take 20 μL system as an example): 10×PCR Buffer (Vazyme, catalog number: P122-d2) 2 μL, dNTP (Vazyme, catalog number: P031-01) 0.2 mM, MgCl 2 1.5mM, Universal Upstream Primer F 0.2μM-0.5μM, Universal Downstream Primer R 0.2μM-0.5μM, Universal Detection Probe P 0.2μM-0.5μM, 50×ROX (Invitrogen, catalog number: 12223-012) 0.4μL, Template 2 μL, make up to 20 μL with sterilized water.
[0094] Control PCR reaction program: 95°C for 3 minutes; 95°C for 15s, 62°C for 30s, 72°C for 30s, 40 cycles.
[0095] Fluorescent signals are detected during the reaction.
[0096] When the specific amplification primer is the dow...
Embodiment 3
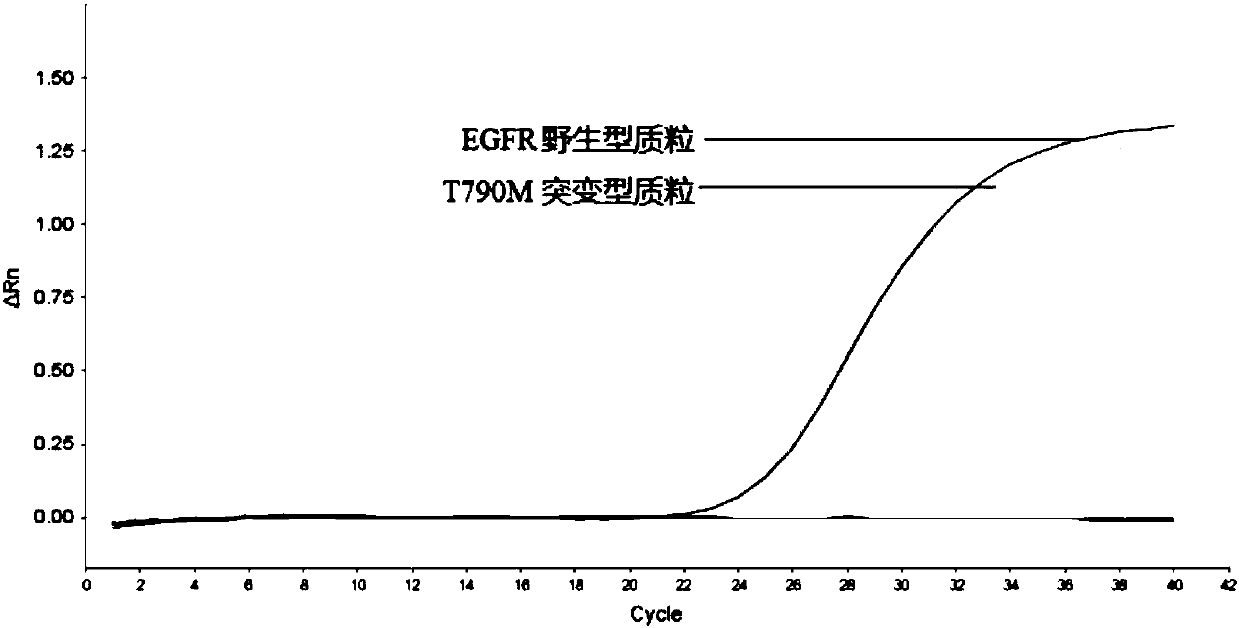
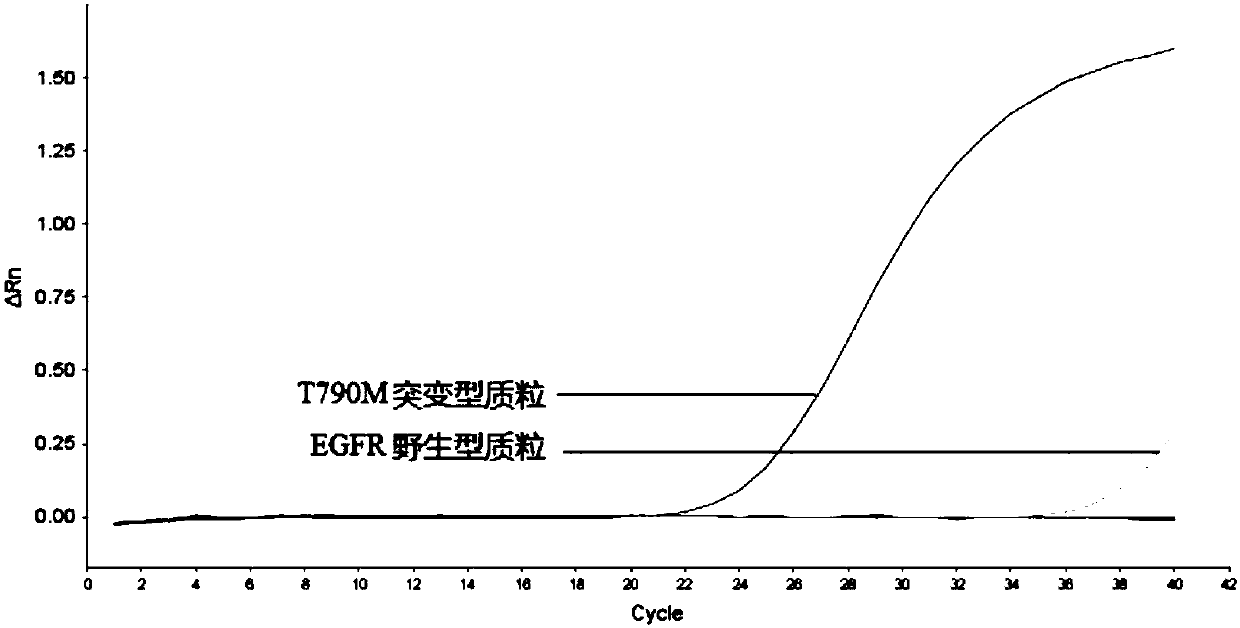
[0102] Example 3, Superblocking Fluorescent Quantitative PCR for Rare Mutation of EGFR Gene T790M
[0103] 1. Detection method
[0104] 1. According to the method of Example 1, primer probes are designed for the EGFR gene T790M mutation (the EGFR gene wild-type target sequence is shown in sequence 6 of the sequence listing; the EGFR gene T790M mutant target sequence is shown in sequence 7 of the sequence listing), As follows:
[0105] Universal upstream primer EGFR-T790M-F: 5'-CCTCCAGGAAGCCTACGTGATGG-3' (SEQ ID NO: 1);
[0106] Universal downstream primer EGFR-T790M-R: 5'-CAGTTGAGCAGGTACTGGGAG-3' (SEQ ID NO: 2);
[0107] Specific amplification downstream primer EGFR-T790M-AP: 5'-AGGG+C+A+TGAGCTGCA-3' (sequence 3); wherein "+" is LNA modification, and "+" indicates subsequent base modification;
[0108] General detection probe EGFR-T790M-P: 5'-TGAGCTGCACGGTGGAGGTGA-3' (SEQ ID NO: 4); wherein the 5' end of the probe is marked with FAM, and the 3' end is marked with BHQ1;
[...
PUM
| Property | Measurement | Unit |
|---|---|---|
| melting point | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com