A self-assembled nucleic acid aptamer/protein composite nanoprobe, preparation method, kit and application thereof
A nucleic acid aptamer and protein complex technology, applied in the field of molecular biology, achieves the effects of low detection limit, improved capture efficiency, high precision and accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
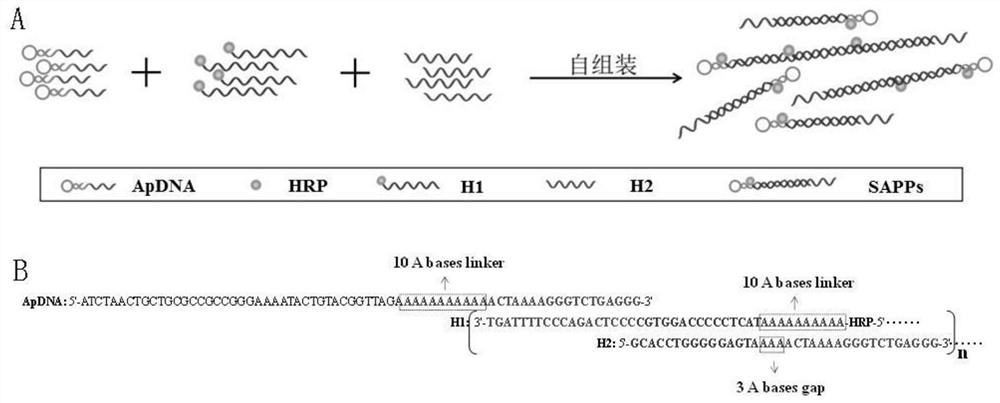
[0057] Preparation of self-assembled nucleic acid aptamer / protein composite nanoprobe: mix 75nmol / L hybridization probe 1, 150nmol / L hybridization probe 2 and 150nmol / L hybridization probe 3 and add to the vortex mixer, vortex at room temperature Spin hybridization for 1 h to obtain the self-assembled nucleic acid aptamer / protein composite nanoprobe. The preparation process is as follows: figure 1 shown.
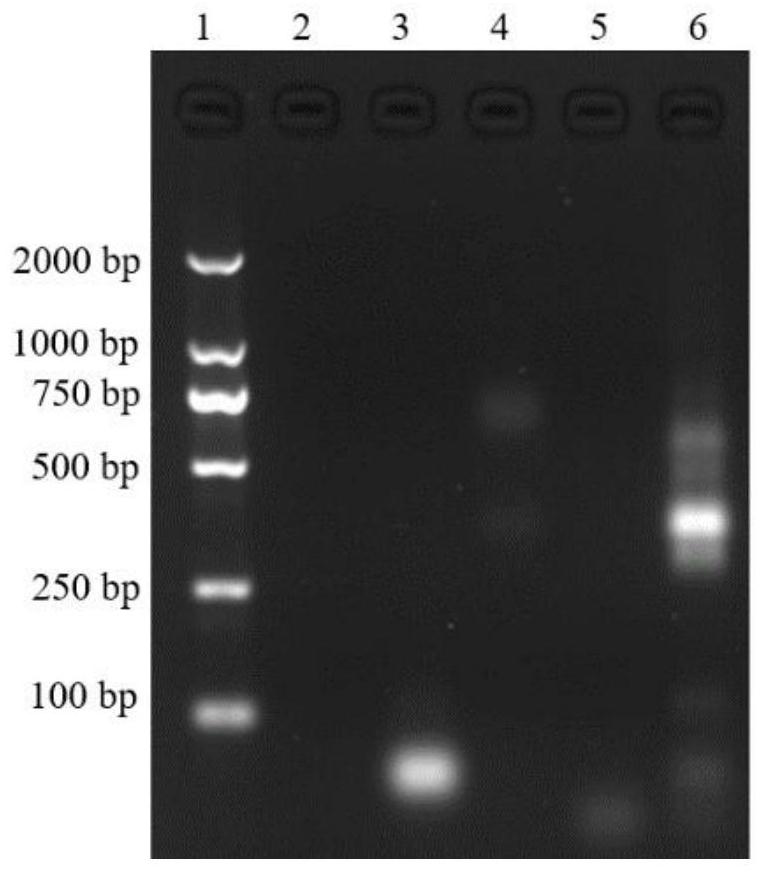
[0058] Characterization of hybridization results: Dilute ApDNA, H1 DNA, and H2 DNA to a concentration of 2 μmol / L, and then take 10 μL of the above solution, 10 μL of a mixed solution of 1 μmol / L ApDNA, 2 μmol / L H1, and 2 μmol / L H2, and perform 2% Agarose gel electrophoresis, the electrophoresis conditions are voltage 100V, time 1h, and finally gel imaging, such as figure 2 shown, where figure 2 Middle lane 1: primer; lane 2: binding buffer; lane 3: 2 μmol / L ApDNA; lane 4: 2 μmol / L H1 DNA; lane 5: 2 μmol / L H2; lane 6: mixed solution of ApDNA, H1 and H2, The concentratio...
Embodiment 2
[0060] The preparation method of Sgc8c-MNPs comprises the following steps:
[0061] A: Wash magnetic beads: Take 300 μL 2mg / mL magnetic beads (MNPs) in a 1.5 mL low adsorption centrifuge tube, magnetically separate, and discard the supernatant. Then add 300 μL of PBS buffer solution, mix thoroughly, magnetically separate, discard the supernatant, and wash 3 times in total.
[0062] B: Combination of MNPs and Sgc8c: Add 500 μL of 400 nmol / L Sgc8c (another portion of Sgc8c with the same concentration, marked as pre) to the above-mentioned low-adsorption centrifuge tube, and mix thoroughly. Vortex reaction at room temperature for 30 minutes, magnetically separate, save the supernatant, marked as post, wash the magnetic beads twice with PBS, 500 μL each time, save the supernatant separately, marked as wash 1 and wash 2;
[0063] C: The MNPs modified with Sgc8c (Sgc8c-MNPs) were resuspended in 600 μL of immunomagnetic bead preservation solution, and stored at 4°C for use.
[0064...
Embodiment 3
[0071] Capture of target cells by Sgc8c-MNPs in a single CCRF-CEM suspension:
[0072] (1) Wash cells: wash CCRF-CEM 3 times with binding buffer, then resuspend for 10 6 cells / mL cell suspension;
[0073] (2) Cell staining: 5 μL of 2 mmol / L DiD dye was added to each ml of CCRF-CEM cell suspension, and incubated at 37° C. for 5 min. Wash cells 3 times with binding buffer;
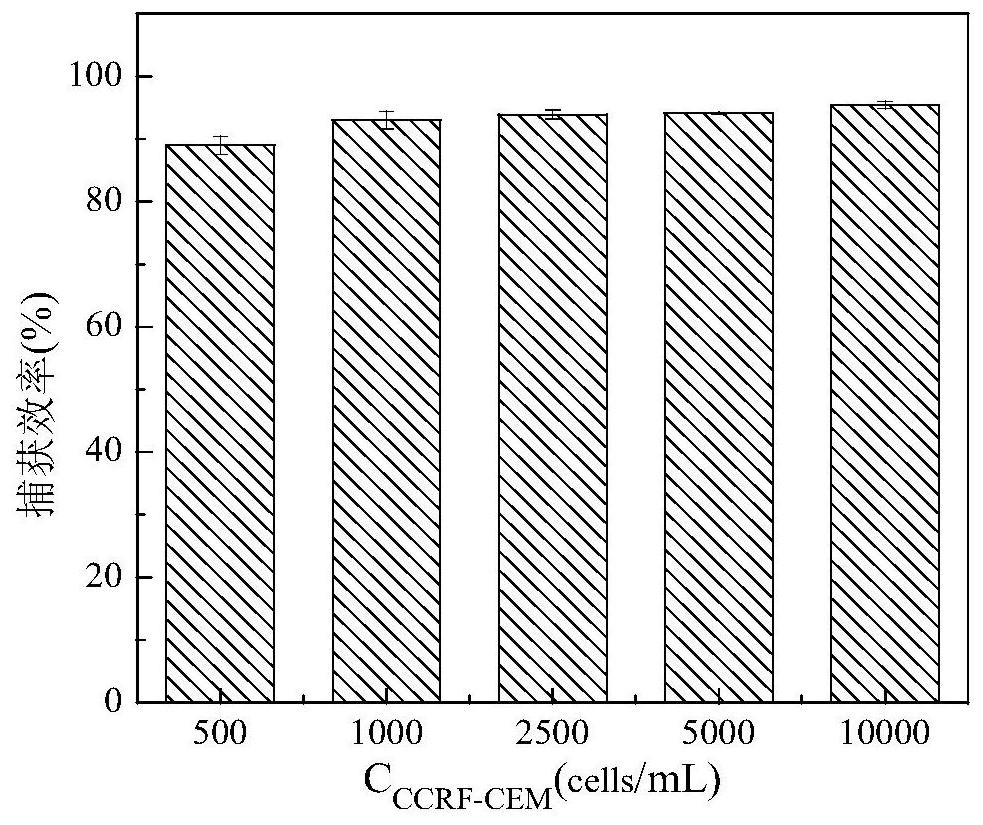
[0074] (3) Sgc8c-MNPs capture CCRF-CEM: Dilute the stained cells into five different concentrations: 500, 1000, 2500, 5000, 10000 cells / mL. Take 1 mL of different concentrations of CCRF-CEM and mix them with Sgc8c-MNPs (the amount of Sgc8c-MNPs is 20 μL of stock solution), incubate at 4°C for 20 min, magnetically separate, and collect the supernatant. Centrifuge at 1000r / min for 5min, and disperse the cell pellet in 100μL binding buffer;
[0075] (4) Cell counting: Under the laser confocal microscope, the CCRF-CEM in the supernatant was counted for fluorescence, and the capture efficiency (Capture effici...
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle size | aaaaa | aaaaa |
| particle size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com