Screening method for reference genes in different tissues of cunninghamia lanceolata and application of screened genes as reference genes
A technology of internal reference genes and screening methods, which is applied in the field of fluorescent quantitative PCR detection, and can solve the problems of screening internal reference genes, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0078] The roots, stems and leaves of Chinese fir seedlings were selected as the research objects of this experiment. The Hoagland nutrient solution was used for sand culture, the pH of the nutrient solution was 5.8, and the formula of the nutrient solution was shown in Table 1. After the cultivation, the roots, stems and leaves of Chinese fir were collected and quickly frozen in liquid nitrogen and stored at -80°C for later use.
[0079] Table 1 Hoagland complete nutrient solution
[0080]
[0081]
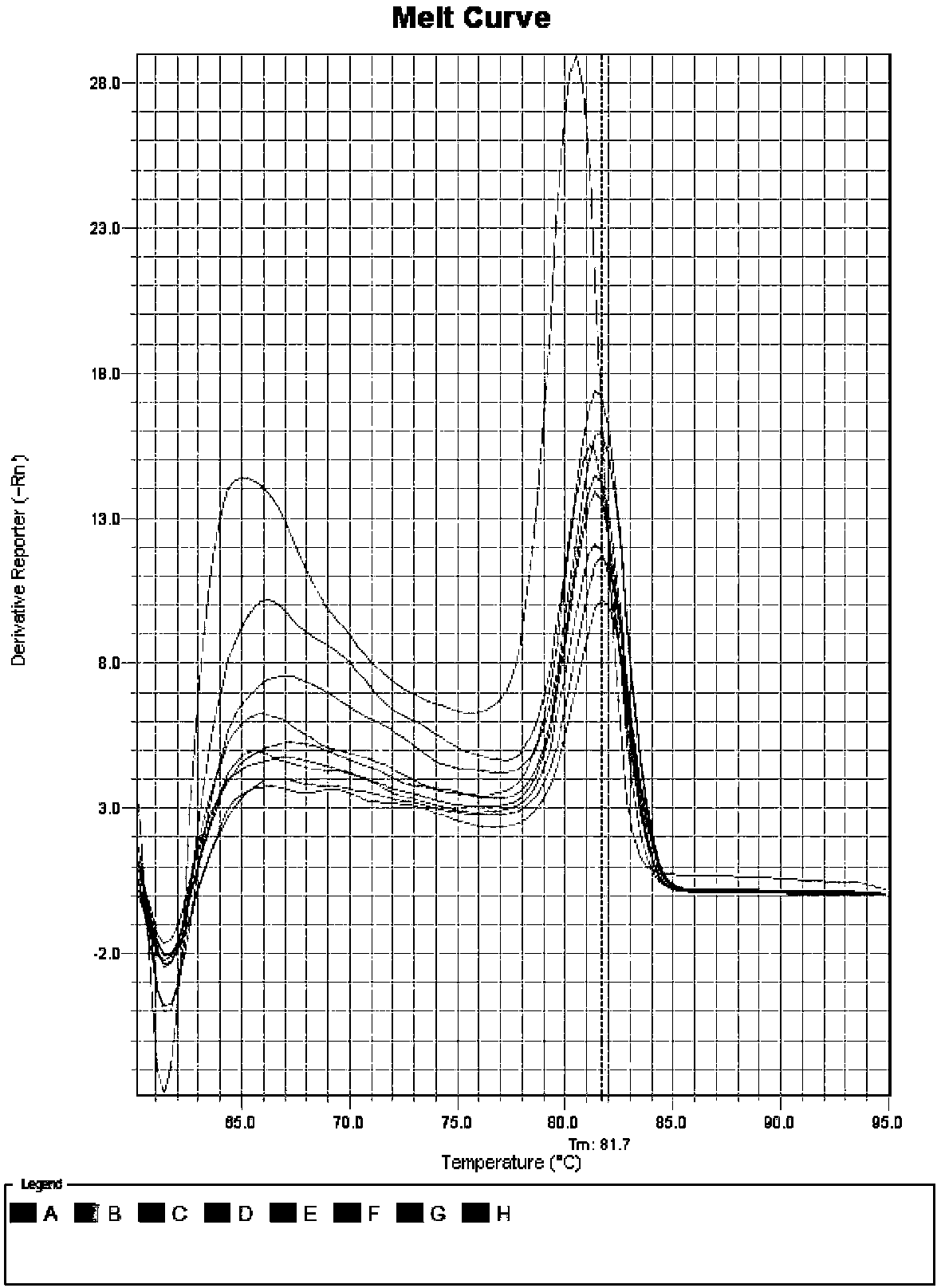
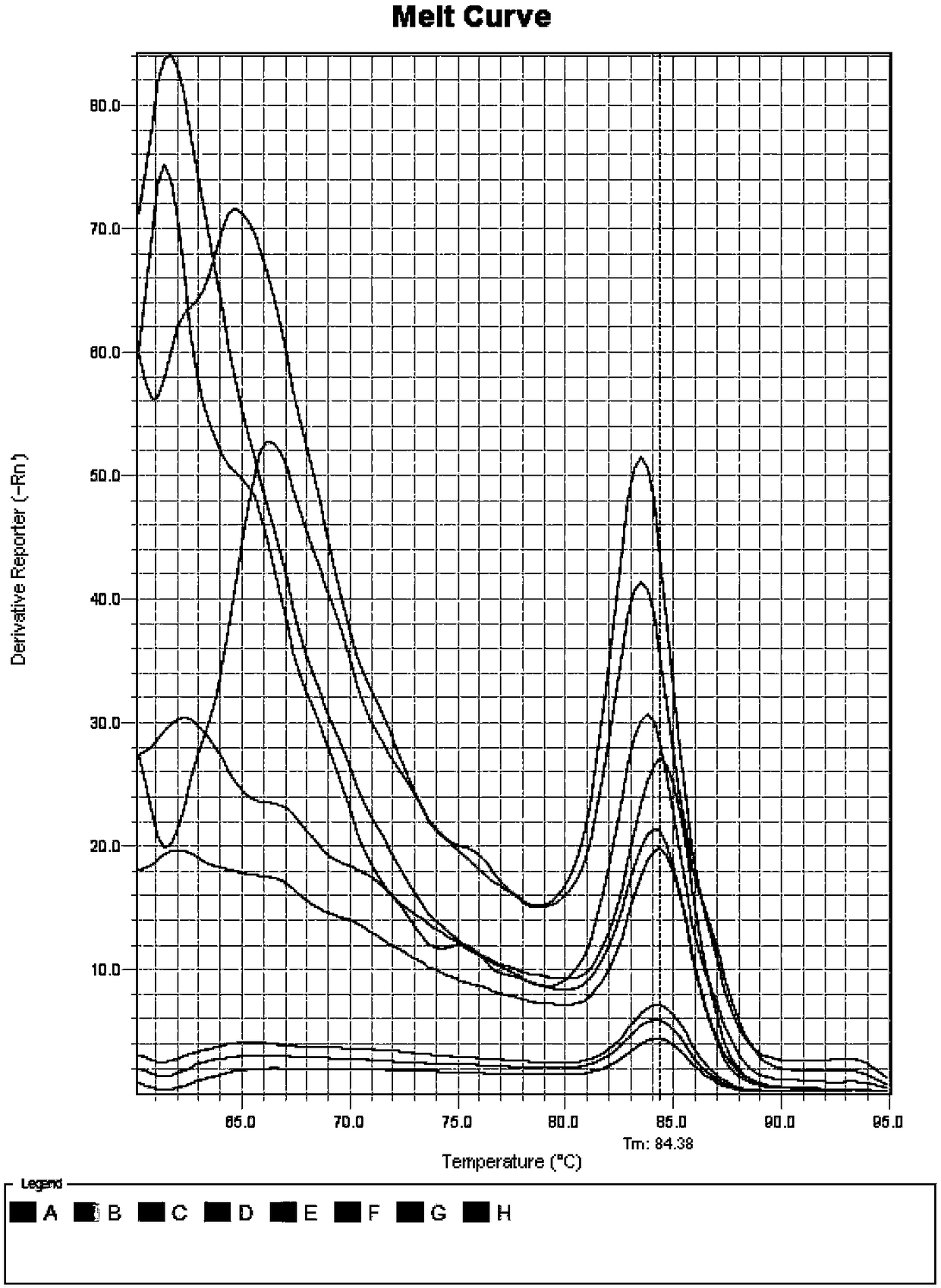
[0082] 1.2 Total RNA extraction and cDNA synthesis
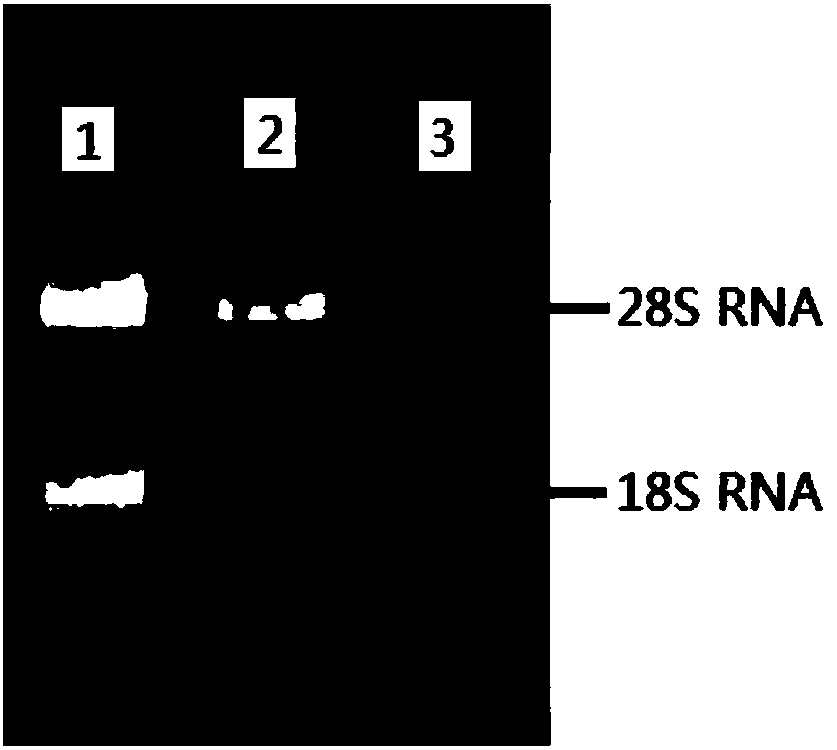
[0083] Based on the RNAprep Pure PlantKit kit from TIANGEN, total RNA was extracted from roots, stems, and leaves of Chinese fir seedlings. The integrity of total RNA was detected by 1.0% agarose gel electrophoresis and stored in a -80°C ultra-low temperature refrigerator.
[0084] The GoScriptTM Reverse Transcription System kit from Promega Company was selected, and a cDNA strand of Chinese fir roots, stems, and leaves ...
Embodiment 2
[0113] Detection of target gene expression
[0114] 18S rRNA was used as an internal reference gene to detect the target gene, which is the transcription factor ClPHR1 of fir phosphorus transporter. Real-time fluorescent quantitative PCR was used to detect the phosphorus supply (1.0mmol / L KH 2 PO 4 ) for 3 days, the expression of ClPHR1 gene in the roots, stems and leaves of No. 32 phosphorus-efficient Chinese fir family. and leaves, with lower expression levels in stems.
[0115] Table 5 Expression analysis of ClPHR1 gene in different tissues of Chinese fir No. 32
[0116] tissue site
Embodiment 3
[0118] 18S rRNA was used as an internal reference gene to detect the expression of Chinese fir cellulose synthase genes ClCesA1 and ClCesA2 in the roots, stems and leaves of No. 32 Chinese fir family. The expression level of ClCesA1 and ClCesA2 is much higher than that in stems and leaves, and the expression level in leaves is lower. The results show that ClCesA1 and ClCesA2 genes are mainly involved in regulating the synthesis of cellulose in Chinese fir stems and roots, which is consistent with the basic functions of these two genes.
[0119] Table 6 Expression analysis of ClCesA1 and ClCesA2 genes in different tissues of Chinese fir No. 32
[0120] tissue site
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com