A SNP marker related to sweet-sour flavor traits of peach fruit and its application
A fruit and acidic technology, which is applied in the direction of recombinant DNA technology, microbial measurement/inspection, DNA/RNA fragments, etc., can solve the problems of high cost, long breeding cycle, low accuracy rate, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
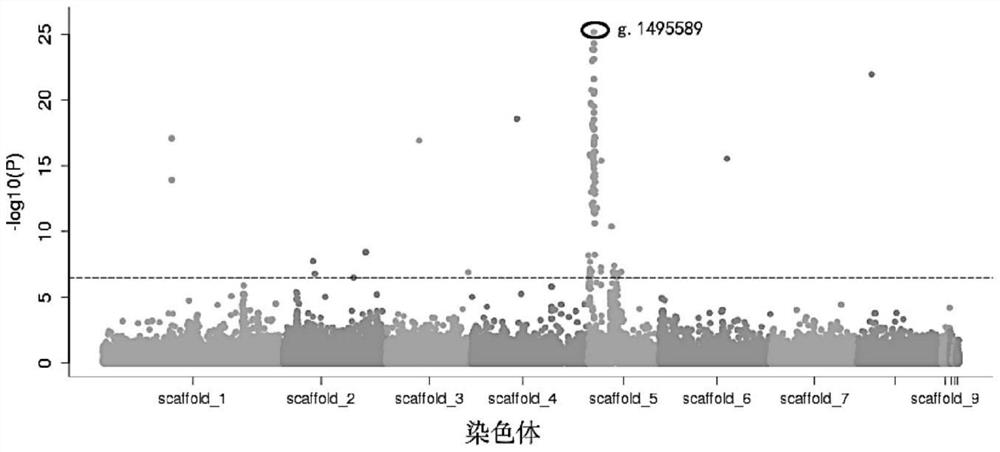
[0038] Example 1. Acquisition of SNP markers related to sweet-sour flavor traits of peach fruit
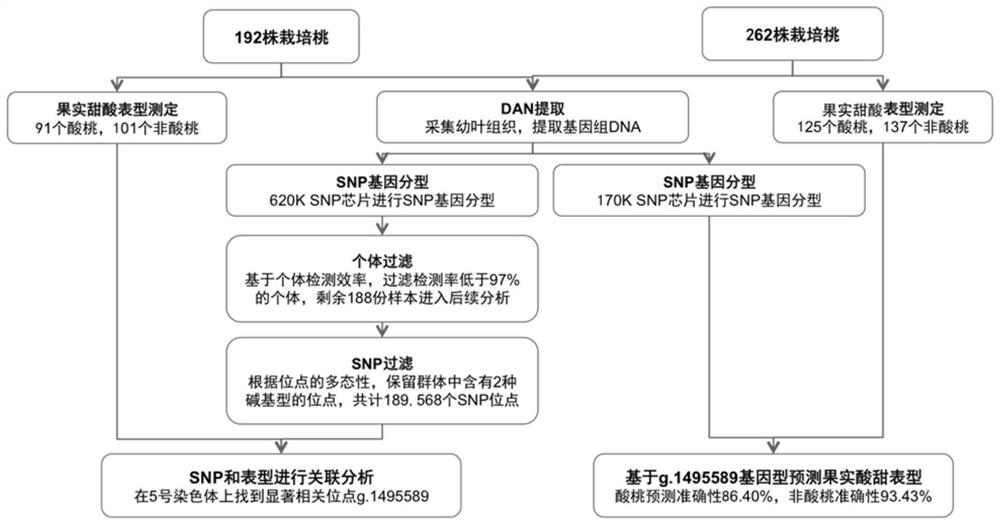
[0039] figure 1 It is the acquisition process of SNP markers related to the sweet-sour flavor traits of peach fruit.
[0040] The experimental materials used in the present invention come from the national fruit tree germplasm Beijing peach and strawberry orchard resource groups.
[0041] In this experiment, a total of 192 peach trees in the resource group were selected, and the phenotypes were identified according to the acid coverage of the fruit surface, including 91 sour peaches (fruit acidic phenotype) and 101 non-sour peaches (fruit non-acidic phenotype) .
[0042] The acidic phenotype of the fruit is that the total acid content of the fruit is greater than or equal to 0.4%, and the pH value of the fruit is less than or equal to 4;
[0043] The non-acidic phenotype of the fruit is that the total acid content of the fruit is less than 0.4%, and the pH value of the fruit is...
Embodiment 2
[0090] Example 2. Application of SNP marker g.1495589 in identifying sweet-sour flavor traits of peach fruit
[0091] 1. Extract the genomic DNA of the sample to be tested
[0092] Collect 262 peach young leaf tissues whose fruit acidic phenotypes and non-acidic phenotypes have been identified, place them in tinfoil, and extract each peach tree genomic DNA, the same as the extraction method in Example 1, to obtain the genome of each sample DNA.
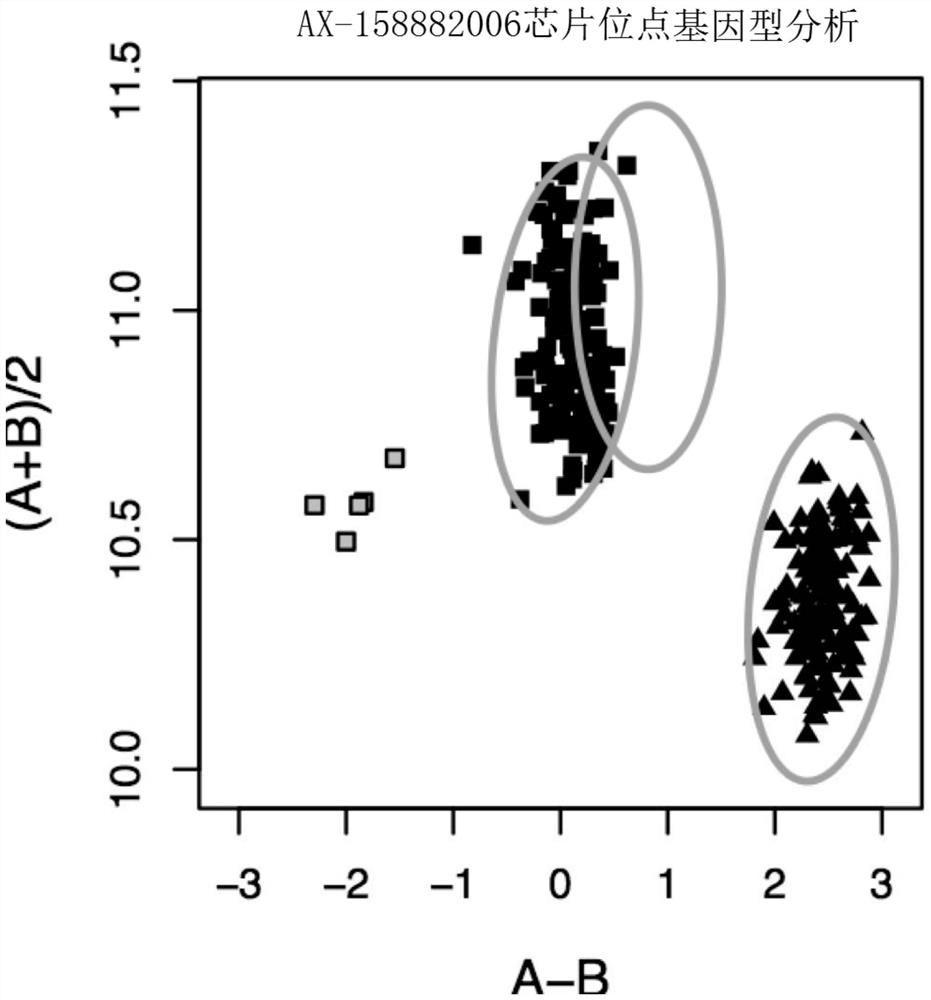
[0093] 2. Genotyping of SNP marker loci
[0094] The genomic DNA of each peach tree mentioned above was used to determine the genotype of the peach genome 170K SNP (Affymetrix, Affymetrix, USA) on the Axiom gene chip of the GeneTitan platform according to the company's standard procedure, and the chip signal was analyzed using the SNPolisher package and Genotyping ConsoleTM of Affymetrix Software (GTC) software package clustering is carried out genotyping, determines the genotype of g.1495589 according to the effect of clustering, a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com