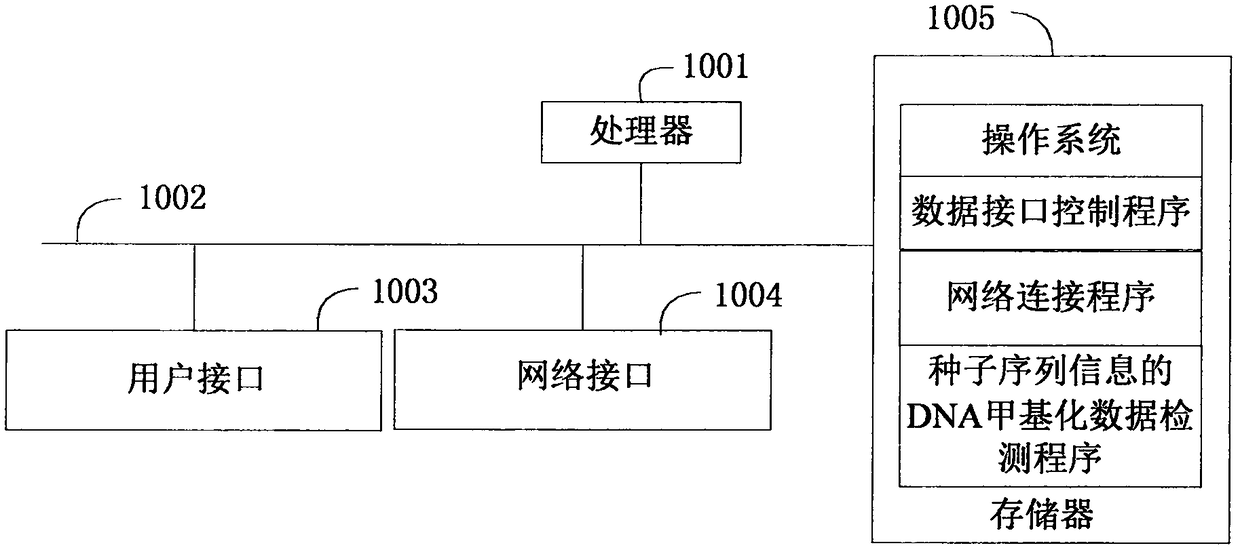
DNA methylation data detection method and device thereof based on seed sequence information
A sequence information and data detection technology, applied in the field of bioinformatics, can solve the problems of low precision, large amount of data, missing sites in the methylation detection region, etc., and achieve improved utilization, convenient research, and shortened comparison operations Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
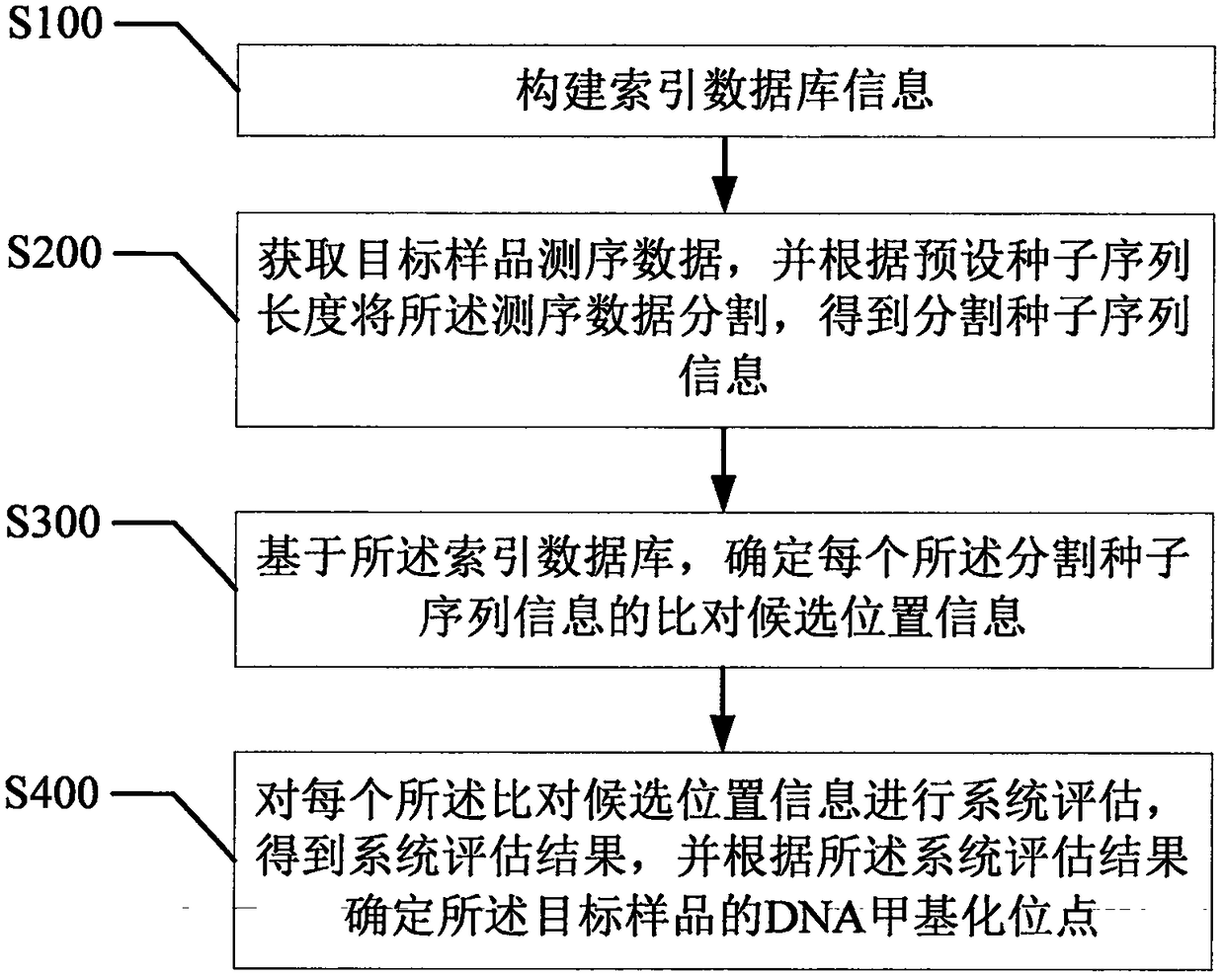
[0071] refer to figure 2 , the first embodiment of the present invention provides a method for detecting DNA methylation data of seed sequence information, comprising:
[0072] Step S100, constructing an index database;
[0073] As mentioned above, in this embodiment, the index database is the reference genome sequence information of the species containing the target sample to be tested. For example, the target sample to be tested is the DNA of the Kunming S180 rat, and then the reference genome sequence information of the relevant rodent is imported. As a comparison reference.
[0074] Step S200, obtaining the sequencing data of the target sample, and segmenting the sequencing data according to the preset seed sequence length to obtain segmented seed sequence information;
[0075] As mentioned above, it needs to be understood that, so far, the bisulfite sequencing method is a relatively efficient method for detecting DNA 5mC base modification, which can directly detect the...
Embodiment 2
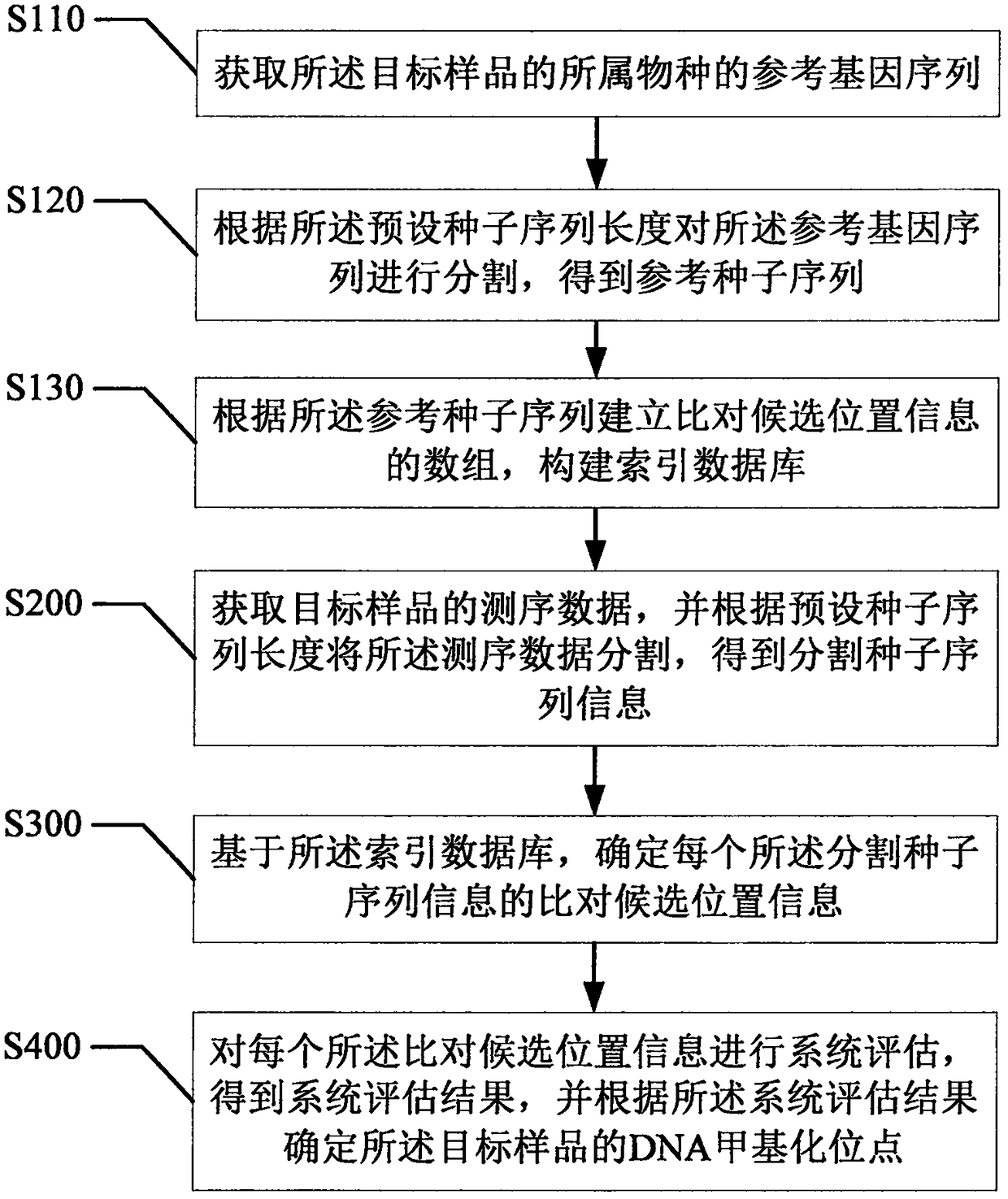
[0084] refer to image 3 , the second embodiment of the present invention provides a DNA methylation data detection method for seed sequence information, based on the above figure 2 In the first embodiment shown, the step S100 includes:
[0085] Step S110, obtaining the reference gene sequence of the species to which the target sample belongs;
[0086] As mentioned above, before performing the methylation mapping of the target sample, it is necessary to construct an index database, and write the reference genome sequence file of the research species corresponding to the target sample into the memory.
[0087] Step S120, segmenting the reference gene sequence according to the length of the preset seed sequence to obtain a reference seed sequence;
[0088] As mentioned above, the reference gene sequence is a long string of four letters representing four bases ATCG. Taking the human reference genome as an example, the actual length is about 3×10 9 bp. The idea of encoding ...
Embodiment 3
[0096] refer to Figure 4 , the third embodiment of the present invention provides a DNA methylation data detection method for seed sequence information, based on the above figure 2 In the first embodiment shown, the step S200 includes:
[0097] Step S210, performing sequencing on the target sample to obtain sequencing data of the target sample;
[0098] As mentioned above, the target sample is sequenced, that is, as Figure 10 As shown, the target sample to be sequenced is treated with bisulfite, and high-throughput sequencing is performed by a next-generation sequencer to obtain the target sample sequencing data. Among them, the sequence information showing DNA will change, the original non-methylated cytosine becomes uracil, and the sequencing result is reflected into thymine, while the methylated cytosine and other bases remain unchanged.
[0099] Step S220, according to the preset seed sequence length, segment the target sample sequencing data segment by segment from ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com