Circular dumbbell-shaped probes and application thereof
A dumbbell-shaped, probe-based technology, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, etc., can solve the problems of reduced cell activity, slow reaction rate, and limited accuracy, so as to reduce false positives and background signals, The effect of improving detection accuracy and precision
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0083] Example 1 A circular dumbbell-shaped probe for detecting target sequences
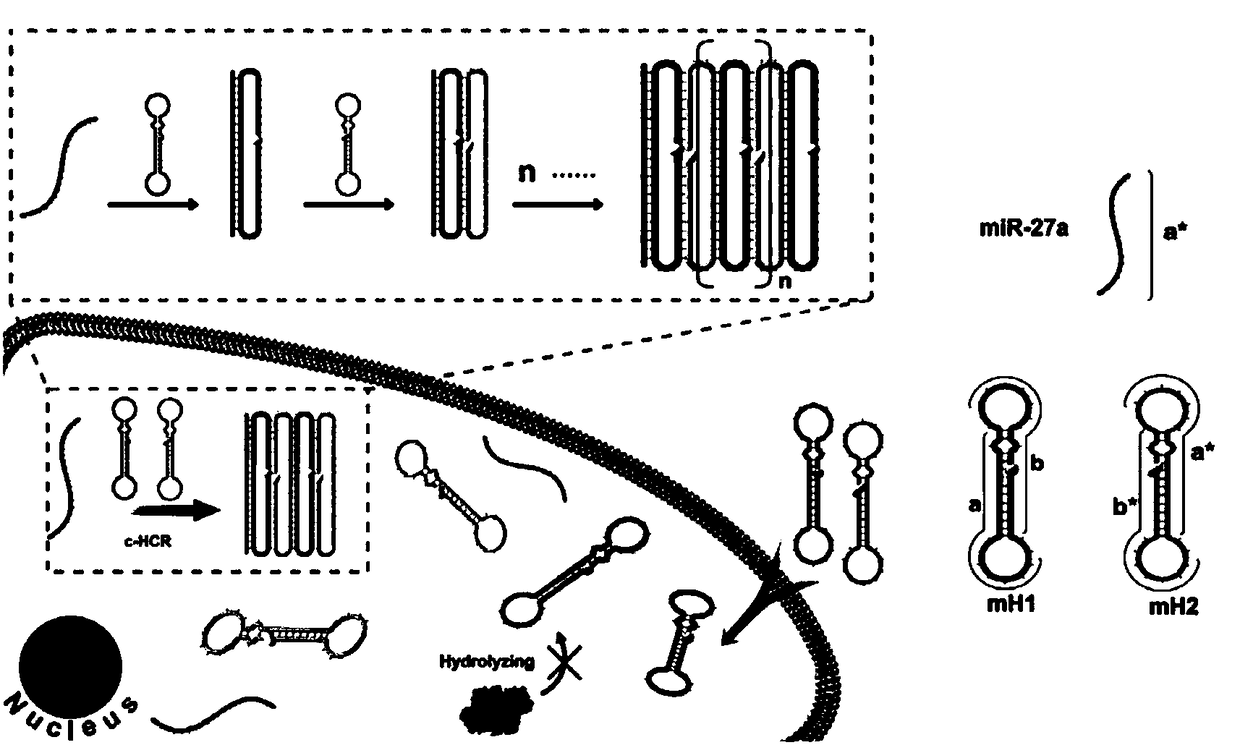
[0084] This embodiment takes the detection of miR-27a: 5'-UUC ACA GUG GCU AAG UUC CGC-3' (SEQ ID NO: 9) in the miR-27 family as an example.
[0085] According to the target sequence miR-27a, two different circular dumbbell-shaped probes were designed, which were respectively denoted as H1 (SEQ ID NO: 1) and H2 (SEQ ID NO: 3). Both H1 and H2 were closed base sequences, Both are dumbbell-shaped, and the rings at both ends are connected together by the neck in the middle, and the neck is composed of reverse complementary paired base pairs;
[0086] H1 contains a fragment X that is reverse-complementary to the target sequence miR-27a. Part of the base sequence (7bp) in X is located at the loop. This part of the sequence is called the toehold sequence, and the remaining sequence (14bp) in X is located at the neck ;
[0087] H2 contains a fragment Y that can perform reverse complementary pairing wit...
Embodiment 2
[0102] Synthesis and detection of embodiment 2 circular dumbbell-shaped probe
[0103] 2.1 Synthesis of circular dumbbell-shaped probes
[0104] Design a series of different types of dumbbell-shaped probes (SEQ ID NO: 1-8), and commission Shanghai BioSynthesis to synthesize them. The 5' end of the probes is modified with a phosphate group, connected by T4DNA ligase and extra-nucleic acid After processing the synthesized probe with Dicer I and Exonuclease III, a purified circular dumbbell-shaped DNA probe can be obtained, and the specific operations are as follows:
[0105] The commissioned probe was denatured at 95°C for 5 minutes, and naturally cooled to room temperature to make the probe an open ring. Take 30 μL of the probe solution, add 5 μL of 10×T4 DNA ligase buffer and 2 μL of T4 DNA ligase (100 U / μL) at 16 °C, react for 2 h, and denature at 65 °C for 10 min to terminate the ligation reaction. Then add exonuclease I (20U / μL) and exonuclease III (100U / μL) to the soluti...
Embodiment 3
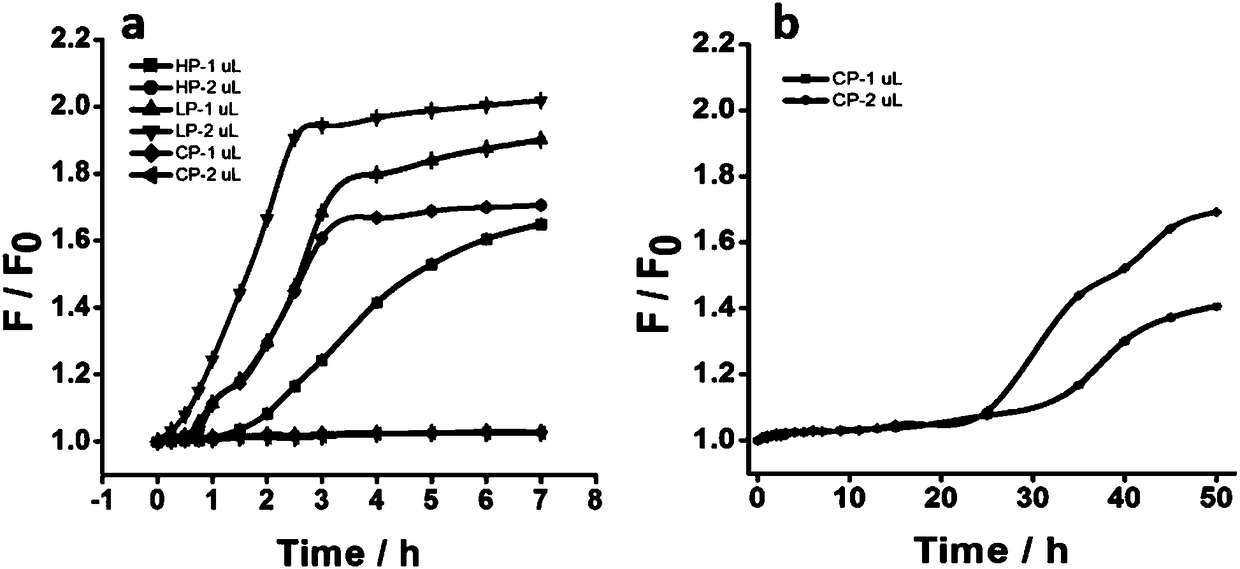
[0108] Embodiment 3 Determination of hydrolysis resistance of ring-shaped dumbbell-shaped probe
[0109] In order to verify the hydrolysis resistance of circular probes, firstly design linear probes L and P containing FAM-BHQ1 labels according to the target sequence miR-27a (L and P are completely complementary, mix L and P in equal proportions to form LP probes) , hairpin probe HP and circular dumbbell-shaped probe CP (H1.2) of the present invention, miR-27a antisense probe block probe. The sequences of each probe are as follows:
[0110] L: 5'-TTCACAGTGGCTAAGTTCCGC(BHQ1)-3' (SEQ ID NO: 10);
[0111] P: 5-(FAM)GCGGAACTTAGCCACTGTGAA-3 (SEQ ID NO: 11);
[0112] HP: 5-(FAM)CGCGCGGAACTTAGCCACTGTGAACGCGCG(BHQ1)-3 (SEQ ID NO: 12);
[0113] Block probe: 5-GCGGAACTTAGCCACTGTGAA-3 (SEQ ID NO: 13);
[0114] CP(H1.2):
[0115] 3-T(FAM)GAATCGGTAAAAGTGTCACCGATT(BHQ1)CAAGGCGCCCACACACCGCCT-5 (SEQ ID NO: 2).
[0116] 3.1 Detection of probe hydrolysis resistance
[0117] The linear pro...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com