Recombinase polymerase amplification (RPA) method, RPA special primers and RPA probe for detecting streptococcus suis serotype 2 as well as application of RPA method, RPA special primers and RPA probe
A Streptococcus suis and probe technology, applied in biochemical equipment and methods, DNA/RNA fragments, recombinant DNA technology, etc., can solve problems such as inability to detect quickly, and achieve the effects of saving detection time, simple method and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Design and screening of embodiment 1, type 2 Streptococcus suis RPA detection primers and probes
[0041] (1) Design of primers and probes
[0042] The inventor analyzed and determined through literature search that the specific sequence in the cps2J gene of Streptococcus suis type 2 used in the present invention is the target gene. The known template gene sequence was obtained from the NCBI database, i.e. the nucleotide sequence shown in SEQ ID NO.1, and the above sequence was synthesized by Nanjing KingScript Biotechnology Co., Ltd. It can be used as a template in the process of needle screening and optimization of reaction system. According to the design principles of RPA primers and probes, 5 primers and 1 probe were designed, as shown in Table 1.
[0043] Table 1 Primer and probe sequences
[0044]
[0045] (2) Primer screening
[0046] Artificially synthesize a positive plasmid containing the sequence shown in SEQ ID NO.1 of the cps2J gene of Streptococcus ...
Embodiment 2
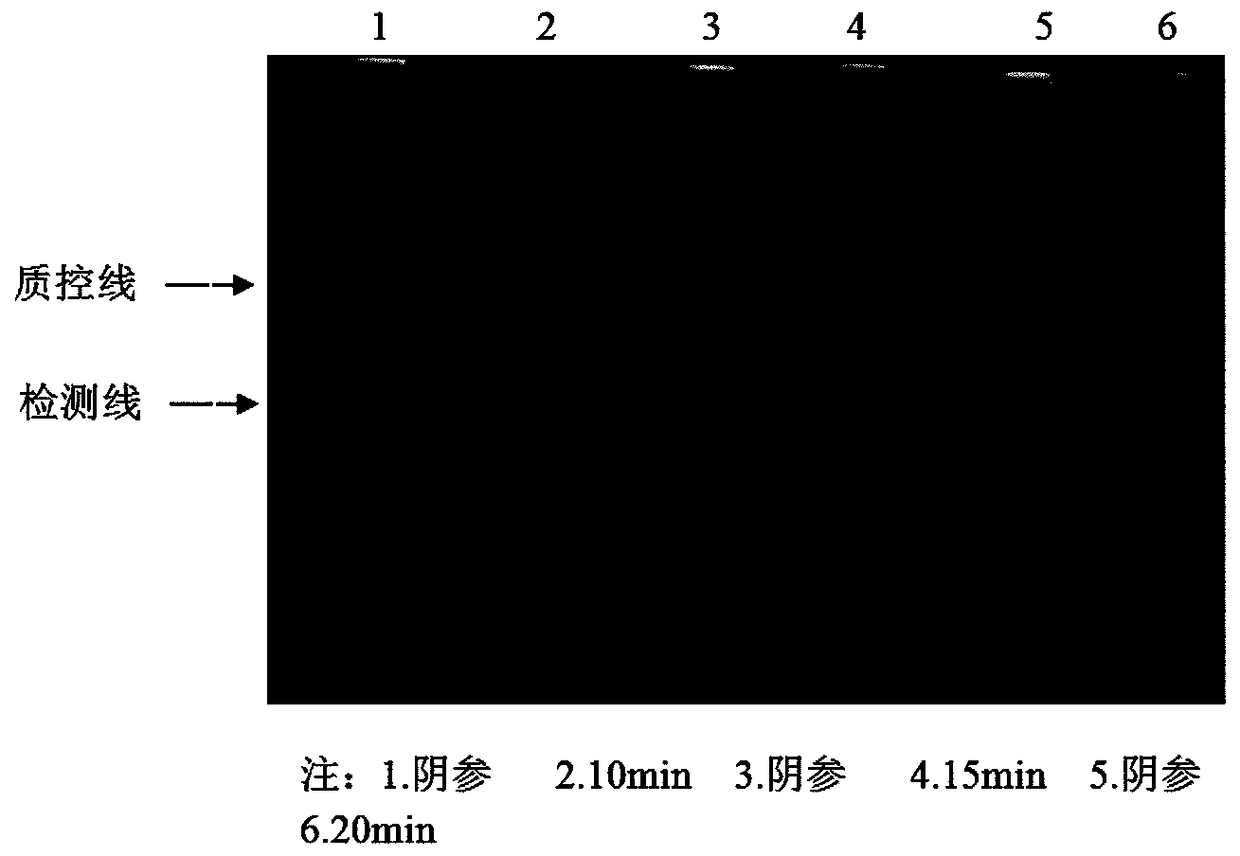
[0055] Embodiment 2: Optimization of RPA reaction system, amplification and detection conditions
[0056] In the process of primer screening, there are still false positives in the detection of lateral flow nucleic acid detection test strips, so the RPA reaction system, amplification and detection conditions need to be optimized
[0057] (1) Primer probe concentration
[0058] Set the concentration gradient of the reverse primer as 10 μmol / L, 5 μmol / L, and 2.5 μmol / L, and the concentration of the probe as 10 μmol / L, 5 μmol / L, and 2.5 μmol / L. The primers were combined with the probes of three concentrations to form 9 combinations, and a set of negative controls was set for each combination. RPA amplification was carried out at 37°C, and the color development of the detection line of the lateral flow nucleic acid detection strip was used as an indicator for the completion of the amplification. The combination with the best amplification effect and no false positives was select...
Embodiment 3
[0067] Embodiment 3: Detection limit evaluation of RPA detection
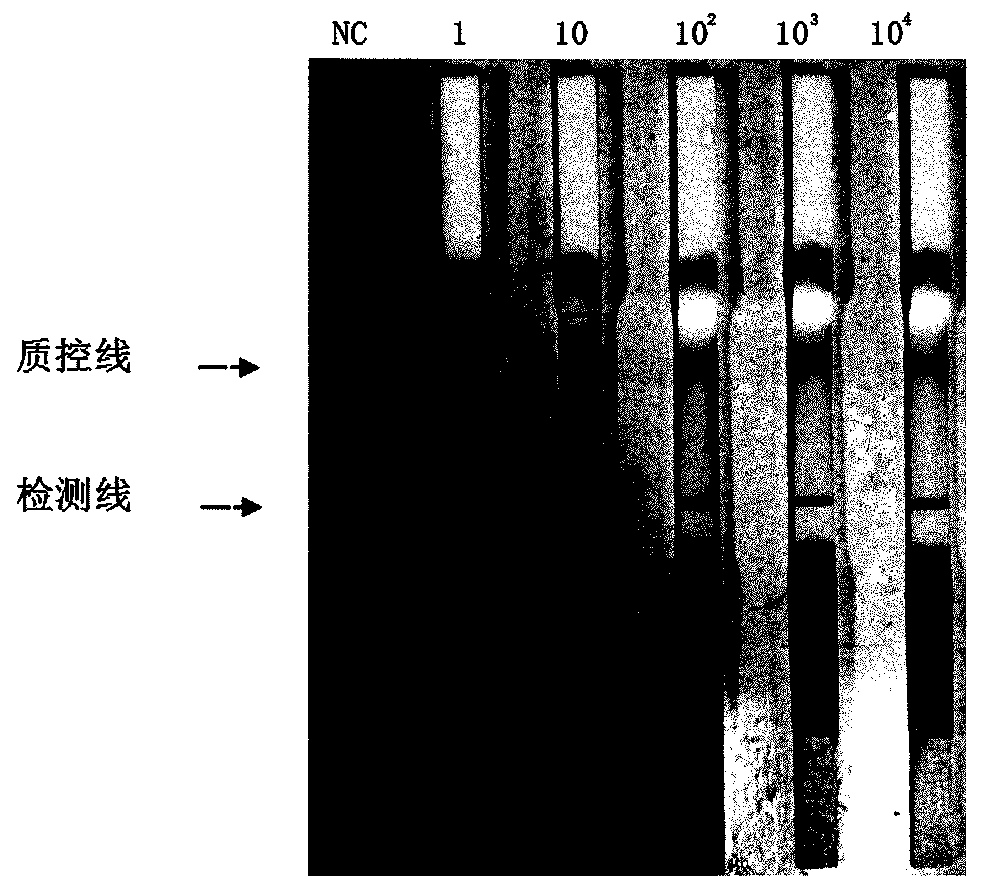
[0068] Dilute the type 2 Streptococcus suis positive plasmid into 10-fold ratio 4 - 10 0 For a series of different concentrations such as copy / μL, 1 μL was added to the reaction system determined in Example 2, and the primer combination screened out was used to perform RPA detection on the templates with different copy numbers above using the reaction conditions determined in Example 2. Observe the detection limit of the RPA assay.
[0069] Results (see Figure 4 ), from 10 2 Copy / μL begins above sample all to be positive, and negative control is negative, illustrates that the detection limit of RPA detection method of the present invention is 10 2 copy / react.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com