A high-throughput sequencing method for detecting multiple variant types of genes and its application
A high-throughput, genetic technology, applied in the field of high-throughput sequencing, can solve problems such as limitations and high detection costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0096] The sample in this example is from a patient with stage IIB breast ductal carcinoma, and the source of the sample is human genomic DNA extracted from an isolated right breast tumor tissue sample.
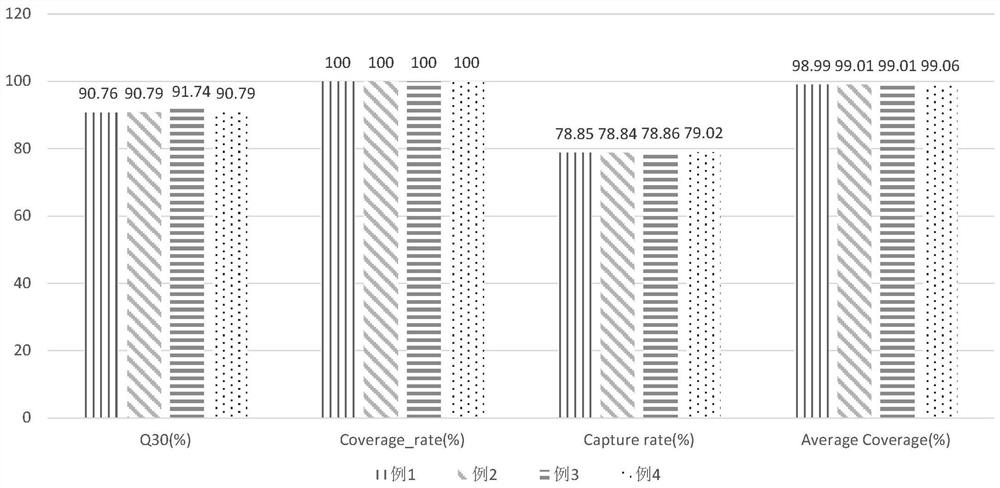
[0097] Complete sample DNA extraction, hybridization and capture of target sequences, elution and enrichment and purification, library quality inspection, on-machine sequencing, and data processing and analysis according to the aforementioned 3-7 steps. The concentration of the obtained DNA library is 27.6ng / μL, and the results are as follows :
[0098] 8. Results
[0099] i) The detection information is as follows:
[0100] Table 4
[0101]
[0102]
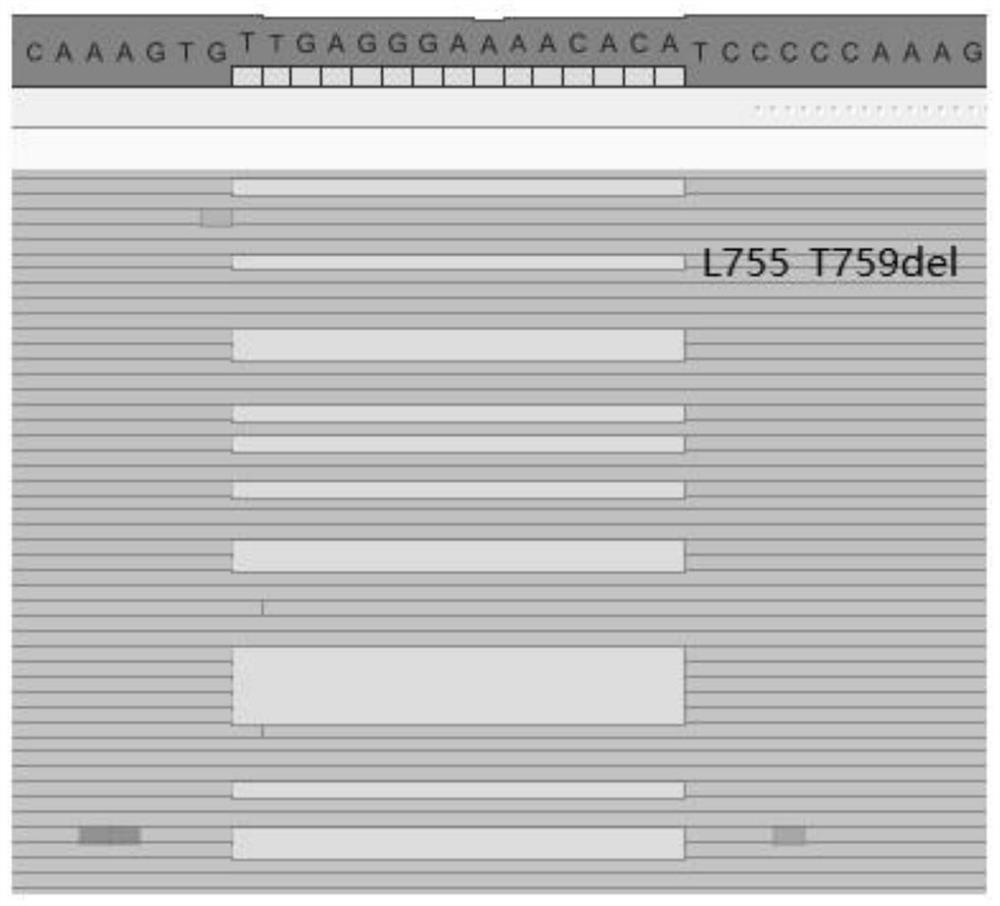
[0103] ii) if figure 2 As shown, the L755_T759del mutation occurred in the ERBB2 gene of this sample, and the sequence data of the sample was compared with the paired normal human genome DNA. The identified mutation site is the L755_T759del (deletion of amino acid 755, 759) mutation in the ERBB2 gene. The mutation is...
Embodiment 2
[0110] The samples in this example are derived from human genomic DNA extracted from tumor tissue samples at the source site in vitro. The tumor was primary triple-negative breast cancer with axillary lymph node metastasis, and the biopsy results reported G3pT1cpN1 ductal type.
[0111] Complete sample DNA extraction, target sequence hybridization capture, elution and enrichment purification, library quality inspection, on-machine sequencing, and data processing and analysis according to the aforementioned 3 to 7 steps. The concentration of the obtained DNA library is 25.8ng / μL, and the results are as follows :
[0112] 8. Results
[0113] i) The detection information is as follows:
[0114] Table 6
[0115]
[0116] ii) if image 3 As shown, the p.T790M mutation occurred in the EGFR gene of the sample, and the sequence data of the sample was compared with the paired normal human genome DNA. The identified mutation site is the p.T790M (790 amino acid change) mutation i...
Embodiment 3
[0123] The sample in this example is derived from isolated left invasive ductal carcinoma of the breast, the pathological stage of the cancer is T2N0M0, and lymphovascular invasion.
[0124] Complete sample DNA extraction, target sequence hybridization capture, elution and enrichment purification, library quality inspection, on-machine sequencing, and data processing and analysis according to the aforementioned 3 to 7 steps. The concentration of the obtained DNA library is 26.2ng / μL, and the results are as follows :
[0125] 8. Results
[0126] i) The detection information is as follows:
[0127] Table 8
[0128]
[0129]
[0130] ii) if Figure 4 As shown, the ERBB2 gene of this sample has a copy number variation, and the sequence data of the sample is compared with the paired normal human genomic DNA. A copy number variation of >4 copies of the ERBB2 gene was found in the sample.
[0131] Table 9
[0132]
[0133] iii). Detailed analysis:
[0134] ERBB2 is a ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com