Method for isolating target cells from a blood sample and use thereof
A blood sample and target cell technology, applied in the field of separating target cells, can solve the problems that need to be strengthened, the detection of specific gene loci is difficult, incomplete, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0092] Example 1 Pretreatment of Peripheral Blood of Pregnant Women and Sorting of Single Cells
[0093] In this embodiment, a pregnant woman sample from a normal male fetus is used, sample number: CFCCYY; male fetus; blood volume: 10 mL; gestational week: 13 weeks.
[0094] Take 10mL of pregnant women's peripheral blood with a heparin tube, add the blood sample to 100mL BD split red solution diluted to 1X in advance, mix gently, and split red at room temperature until the blood is clear, centrifuge to absorb the supernatant, Gently pipette the cell pellet, wash twice with PBS containing 2% FBS, resuspend the cell pellet in 80 microliters of PBS containing 2% FBS, and blow evenly. Add CD45 antibody magnetic beads, incubate at 4°C for 15 minutes, centrifuge at 300g for 10 minutes, discard the supernatant, and resuspend the pellet in PBS containing 2% FBS. Assemble a magnetic bead sorting system (Miltenyi), collect CD45-negative outflow cells, centrifuge to remove the supernata...
Embodiment 2
[0095] Whole genome amplification of embodiment 2 single cell
[0096] The single cells obtained in Example 1 were subjected to whole genome amplification, as follows:
[0097] 1. Preparation of kits for single-cell whole-genome amplification
[0098] The kit consists of cell denaturation lysate, neutralization buffer and amplification system.
[0099] Cell denaturation lysate: composed of solute and solvent; solvent is water; solute and its concentration are as follows: 0.1mM KOH, 1mM EDTA and 0.1M DTT.
[0100] Neutralization buffer: composed of solute and solvent; solvent is water; solute and its concentration are as follows: 60mM KH 2 PO 4 and 5mMK 2 HPO 4 .
[0101] The amplification system includes: 10×Phi29buffer, water, 1.8-2.2×10 -8 mol dNTP, total concentration of primers is 1.8-2.2×10 -11 mol N8primers, 4.5-5.5×10 -3 mg BSA, 0.0045-0.0055 μL Pluronic F68 and 9-11UPhi29 DNA polymerase. Specifically, taking 40.3 μL as an example, the amplification system: 30...
Embodiment 3
[0113] Example 3 Q-PCR detection and STR site detection of single cells
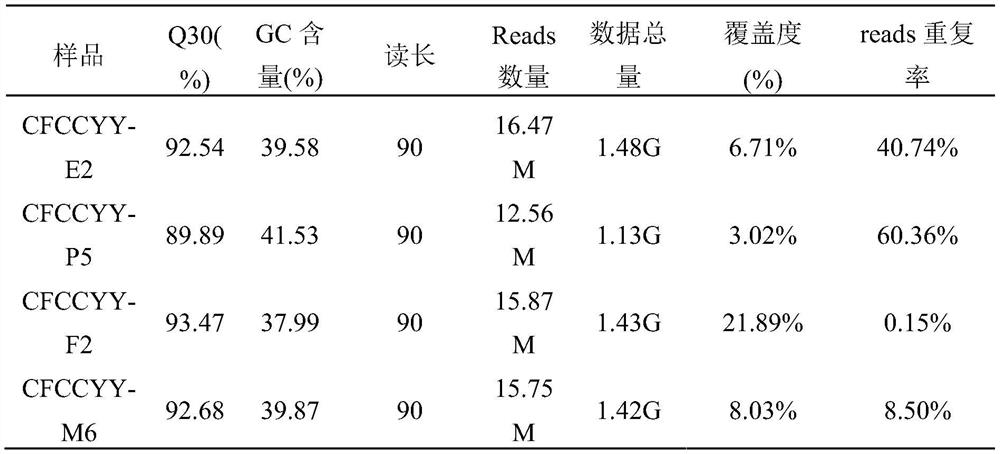
[0114] The 114 single-cell amplification products obtained from the above amplification were used Trio DNAQuantification Kit (Applied Biosystems TM ) for Q-PCR detection. The instrument adopts ViiA 7 real-time fluorescent quantitative PCR instrument, and the reaction conditions of amplification refer to the procedures provided by the kit. Among the 114 products, the amplification products of 4 cells had obvious Y amplification signal.
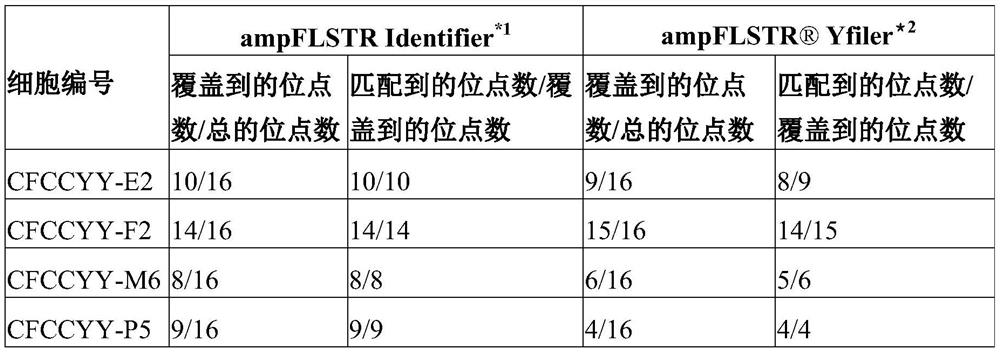
[0115] The inventor further used the ampFLSTRIdentifier PCR amplification kit and the genomes of these 4 single-cell amplification products and parents The Yfiler amplification kit was used for STR detection, and the results are shown in Table 1:
[0116] Table 1: Conventional STR and Y chromosome STR loci
[0117]
[0118] Note: * 1: The ampFLSTR Identifier PCR Amplification Kit can simultaneously amplify and detect 16 STR sites at the same time, so the total ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com