Novel labeling method for messenger RNA and circular RNA based on bimolecular fluorescence complementation
A bimolecular and messenger technology, applied in the field of cell biology, can solve problems such as difficult sequence distinctions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
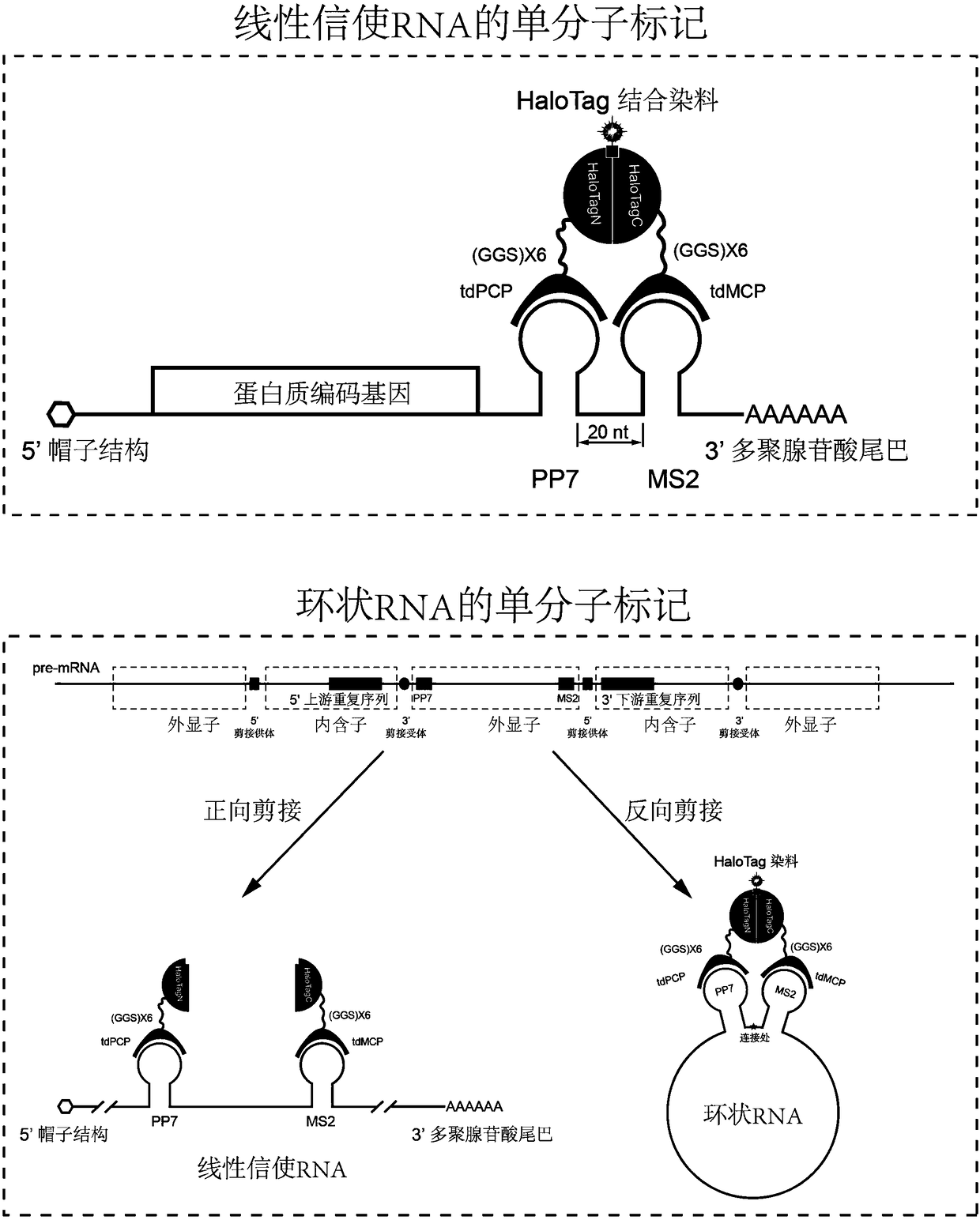
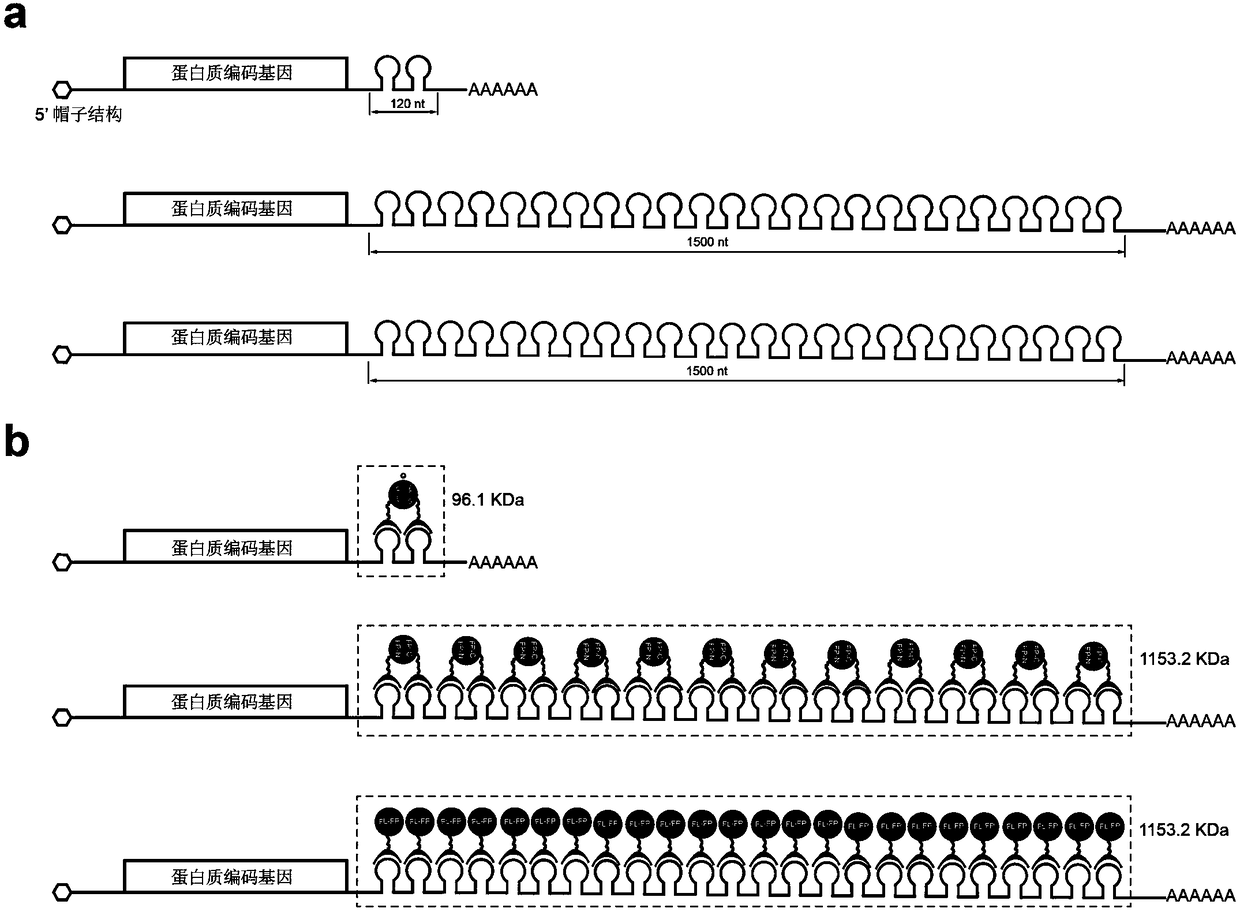
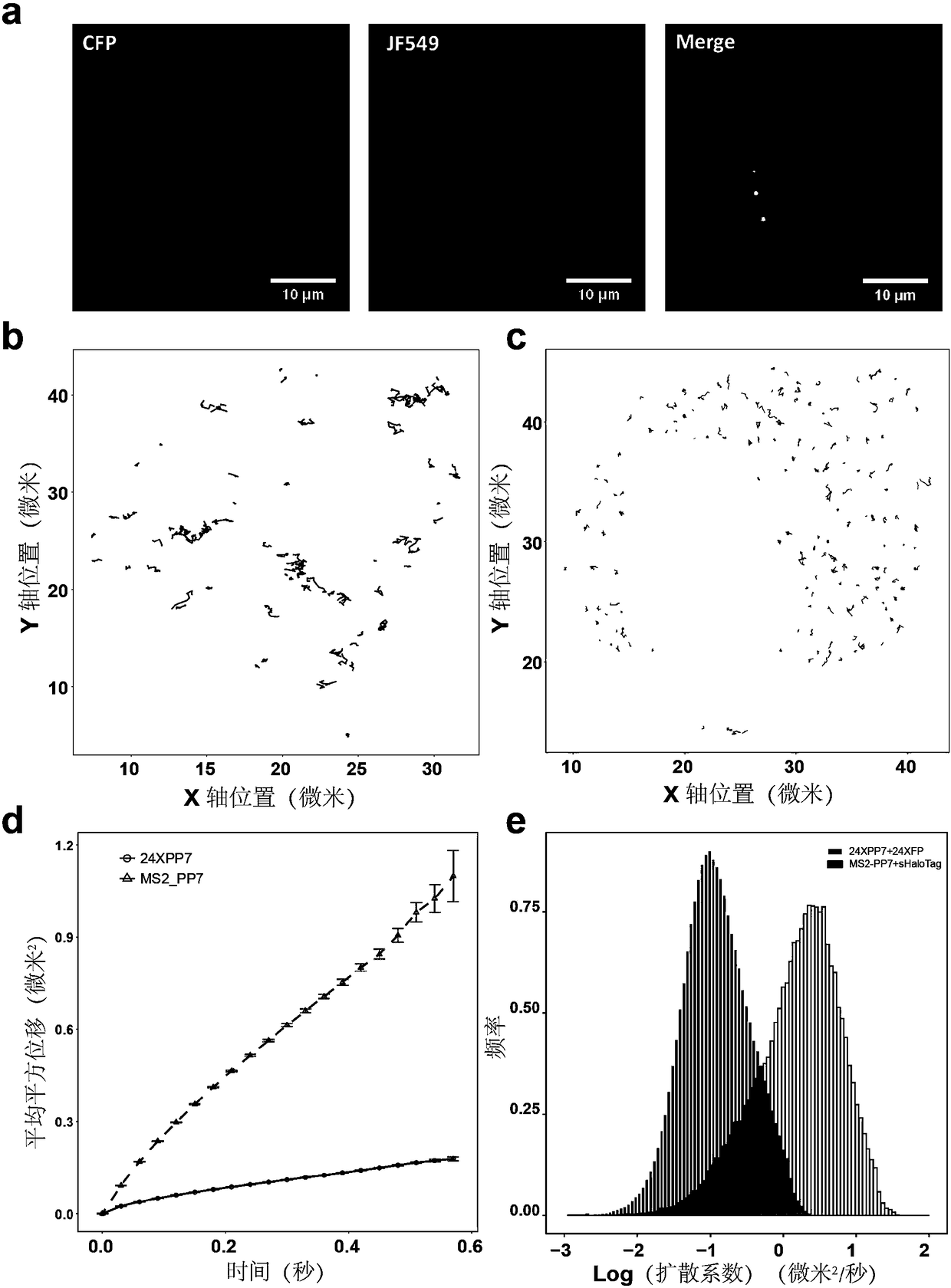
[0024] Example 1: Bimolecular fluorescent complementary labeling of intracellular messenger RNA based on self-ligation tags. Take the messenger RNA of the split HaloTag to mark CFP as an example:
[0025] (1) The forward and reverse strands of MS2 and PP7 were directly synthesized, mixed in equal proportions, annealed, and connected to the 3' end of the non-coding region of the CFP gene to obtain the vector pcDNA3.1(+)-CFP-MS2-PP7.
[0026] (2) The corresponding DNA fragments were amplified from the plasmids containing tdMCP and tdPCP by PCR. Add restriction enzyme sites to the amplified primers. Purified DNA fragments were recovered by DNA agarose gel electrophoresis and a gel purification kit to obtain tdMCP and tdPCP fragments. The PCR amplification system used is as follows:
[0027]
[0028] (3) The tdMCP and tdPCP recovered in the previous step and the eukaryotic expression vector pcDNA3.1(+) were digested overnight with restriction enzymes respectively, and purifi...
Embodiment 2
[0039] Example 2: Bimolecular fluorescent complementary labeling of intracellular circular RNA based on self-ligation tags. Take the circular RNA of ZKSCAN1(548-1047) marked by the split HaloTag used as an example:
[0040] (1) Directly synthesize the positive and negative strands of MS2 and PP7 sequences, mix them in equal proportions, anneal, and connect them into ZKSCAN1 (548-1047) between the splicing donor and acceptor to obtain the vector pcDNA3.1(+)-CircRNA Mini Vector- MS2-ZKSCAN1(548-1047)-PP7.
[0041] (2) The human breast cancer cell line MDA-MB-231 was digested with trypsin, replanted in a 35mm Petri-Dish dedicated for imaging at a density of 50-60%, and cultured for 24 hours.
[0042] (3) The above cells were co-transfected with 1 μg of vectors pcDNA3.1(+)-CircRNAMini Vector-MS2-ZKSCAN1(548-1047)-PP7 and pcDNA3.1(+)-HaloTag N58 using 6mL liposome Lipo-2000 -tdPCP, and pcDNA3.1(+)-tdMCP-HaloTag C58. The medium used for transfection was serum-free Opti-MEM. After ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com