Real-time fluorescent quantitative PCR (polymerase chain reaction) primer, kit and method for detecting virus titer of slow viruses
A real-time fluorescence quantification and virus titer technology, applied in the field of kits and real-time fluorescence quantitative PCR primers, can solve problems such as inability to accurately measure virus titers, inaccurate virus titer detection results, and inability to reflect the true infection activity of the virus.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] The design of embodiment 1 specific primer
[0040] According to the published virus sequence, the present invention selects the envelope region of lentivirus to design primers; conducts preliminary screening on the designed primers to obtain a set of primer pairs with high versatility, good sensitivity and specificity, Its nucleotide sequence is as follows:
[0041] ENV-F:CGCAGTTAATCCTGGCCTGTTA (SEQ.ID:NO.1);
[0042] ENV-R: ATCCTTTGATGCACACAATAGAGG (SEQ. ID: NO. 2).
[0043] Using the lentiviral shuttle plasmid as a template, the amplified sequence is:
[0044] cgcagttaatcctggcctgttagaaacatcagaaggctgtagacaaatactgggacagctacaaccatcccttcagacaggatcagaagaacttagatcatttataatacagtagcaaccctctattgtgtgcatcaaaggat (SEQ.ID:NO.3);
[0045] The length of the amplified fragment is 144bp.
[0046] Internal reference BMP2 (Homo sapiens bone morphogenetic protein 2, bone morphogenetic protein 2) primers:
[0047] BMP2-F:TAGGGTAGACAGAGCCAAGG (SEQ.ID:NO.4);
[0048] BMP2-R: AGCACAGG...
Embodiment 2
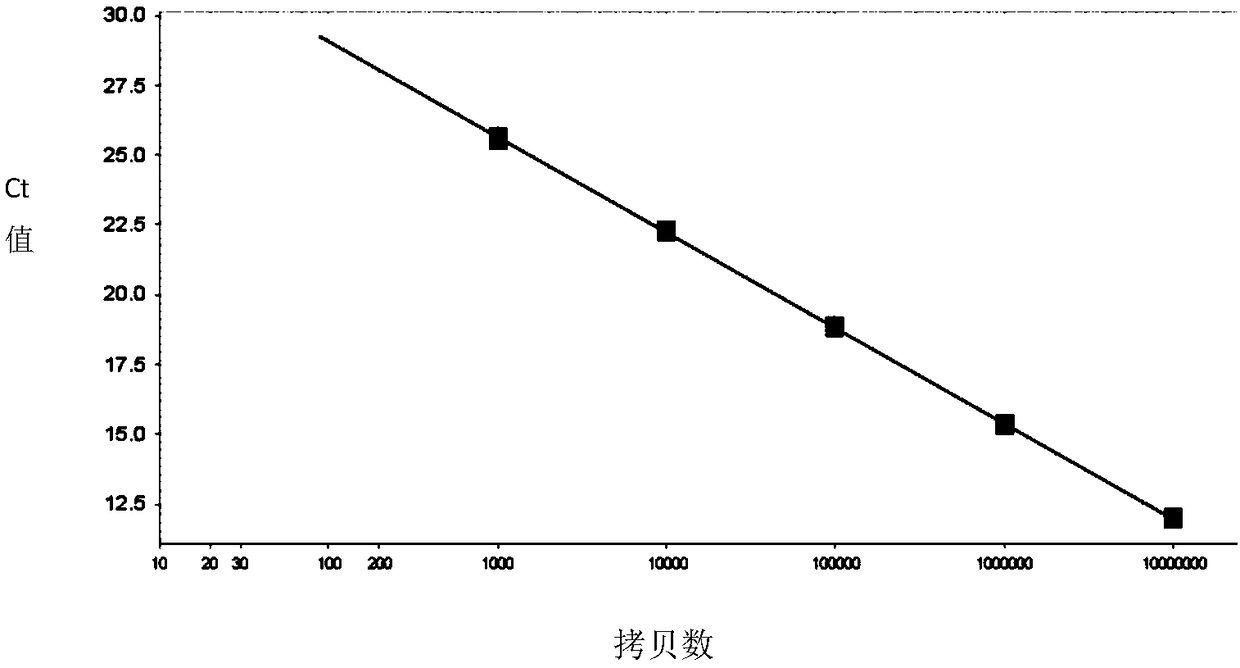
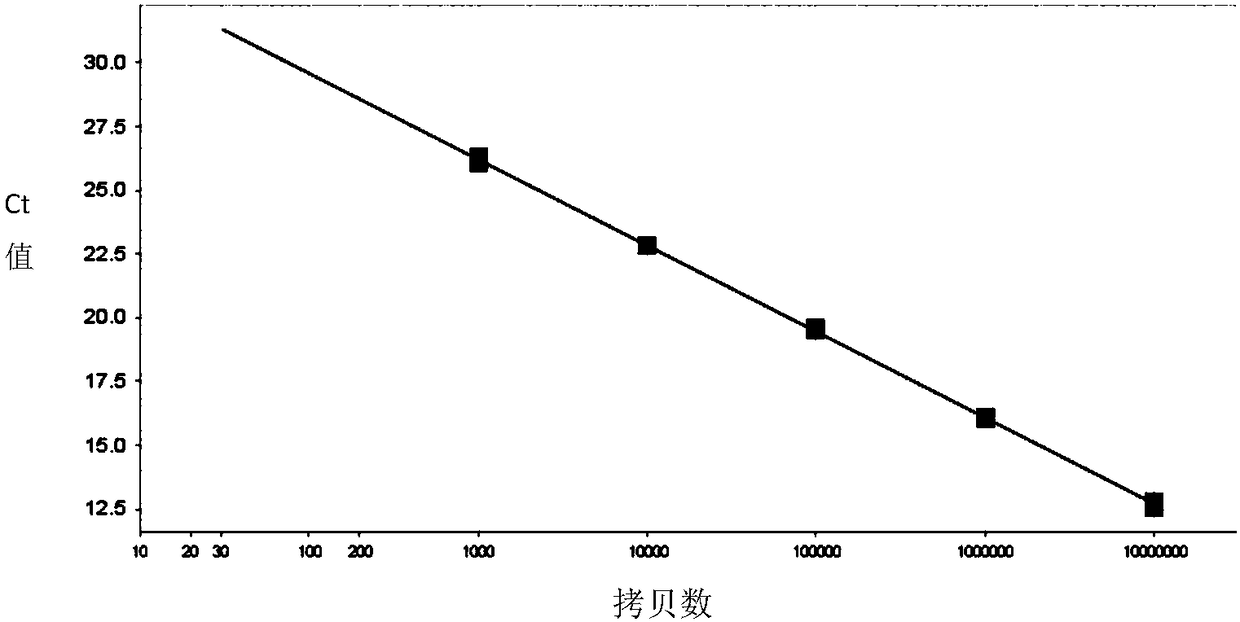
[0052] Preparation and real-time quantitative PCR (qPCR) detection of embodiment 2 standard
[0053] 1. Preparation of Standards
[0054] Use the ENV-F, ENV-R, BMP2-F, BMP2-R primers of Example 1 to amplify the target fragment, and connect the amplified product to pMD TM 18-T vector. The constructed vector was double digested with AhdI and NdeI, and the digested product was recovered and its concentration and purity were determined using NanoDrop. For samples that meet the purity, the number of molecules is calculated according to the molecular weight of the fragment and Avogadro's constant, and used as standards for virus quantification, which are ENV standards and internal reference BMP2 standards.
[0055] The specific calculation formula is: sample copy number per 1uL fragment = A×N×10 -9 / M. Wherein A is the measured DNA concentration in ng / uL; N is Avogadro's constant (6.02×10 23 / mol); M is the molecular weight of the fragment.
[0056] 2. Real-time quantitative PCR...
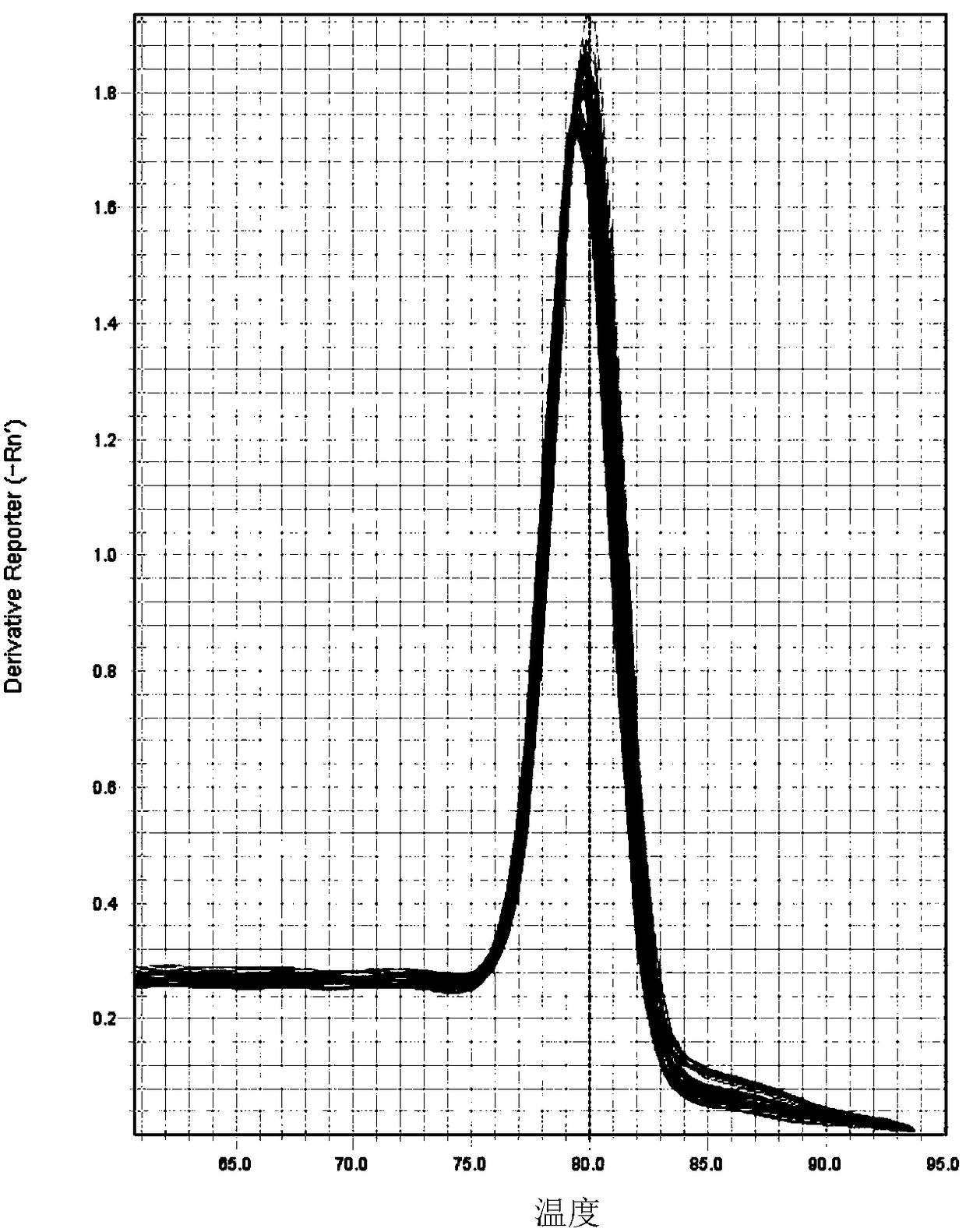
Embodiment 3
[0068] Example 3 Real-time fluorescent quantitative PCR of target cell genomic DNA
[0069] (1) Virus transduction of 293T cells:
[0070] 3 x 10 per well 5 Inoculate 293T cells into a 6-well plate, take a well of cells before virus transduction the next day, trypsinize and count the cells, and save the counting results as B. Use 293T complete medium to dilute the virus product 100 times, take 50uL of the diluted virus solution and add it to one well of a 6-well plate, and add 10uL, 0.5mg / mL polybrene (polybrene) to promote transduction, and keep one The cells that were not transduced with any virus were used as a negative control and placed in a 37-degree incubator to continue culturing.
[0071] (2) Extraction of cellular genomic DNA:
[0072] After 48 hours of virus transduction, the cells were collected, and the genomic DNA of the target cells was extracted with a blood tissue cell genome extraction kit (Tiangen, DP304).
[0073] (3) Quantitative PCR analysis:
[0074...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com