Prostatic cancer stem cell marker, application of antibody OV6 in preparation of prostatic cancer stem cell marking material and marking method
A technology for stem cell markers and prostate cancer, which is applied in the field of preparation of prostate cancer stem cell marker materials, can solve problems affecting the exploration of prostate cancer stem cells in depth and clinical application, and achieve enhanced labeling effect, important clinical value, and high yield. tumor rate effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Source of cell lines: Prostate cancer cell lines C4-2 and C4-2B were provided by Dr. Leland Chung certified (Cedars-Sinai Medical Center, Los Angeles, CA, USA). C4-2 and C4-2B cells were cultured in RPMI 1640 medium (11835093, Gibco, USA) containing 10% fetal bovine serum (FBS, 10099-141, Gibco, USA), supplemented with 1% penicillin / streptomycin ( 15140122, Gibco, USA).
[0039] Enzalutamide (Enzalutamide, ENZ, S1250, Selleck, USA)
[0040] ① Divide 5×10 6 C4-2 and C4-2B cells were plated in a 10 cm dish (430167, Coring, USA), and enzalutamide (5 nM) was added after 24 hours, and stimulated for 48 hours.
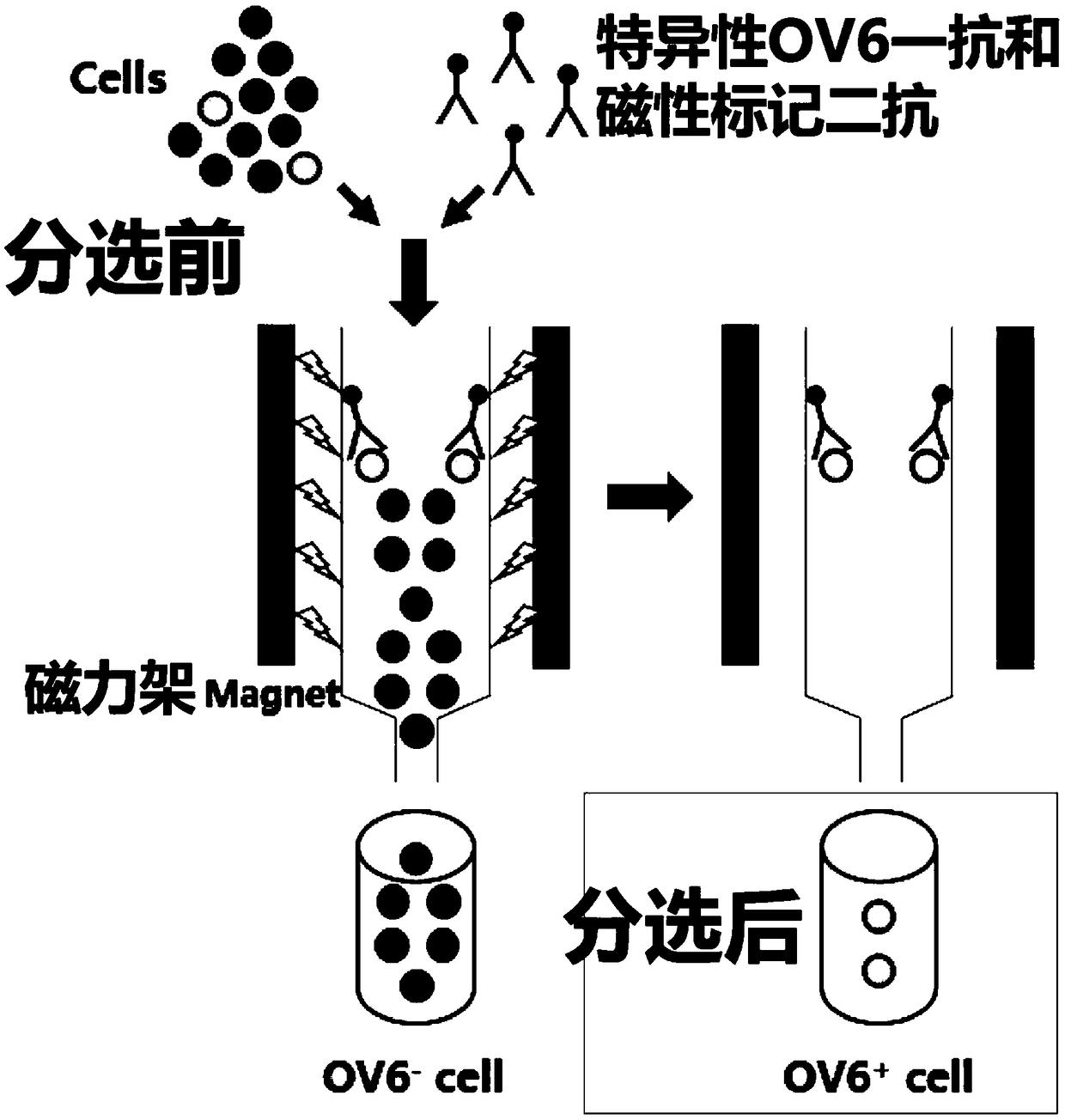
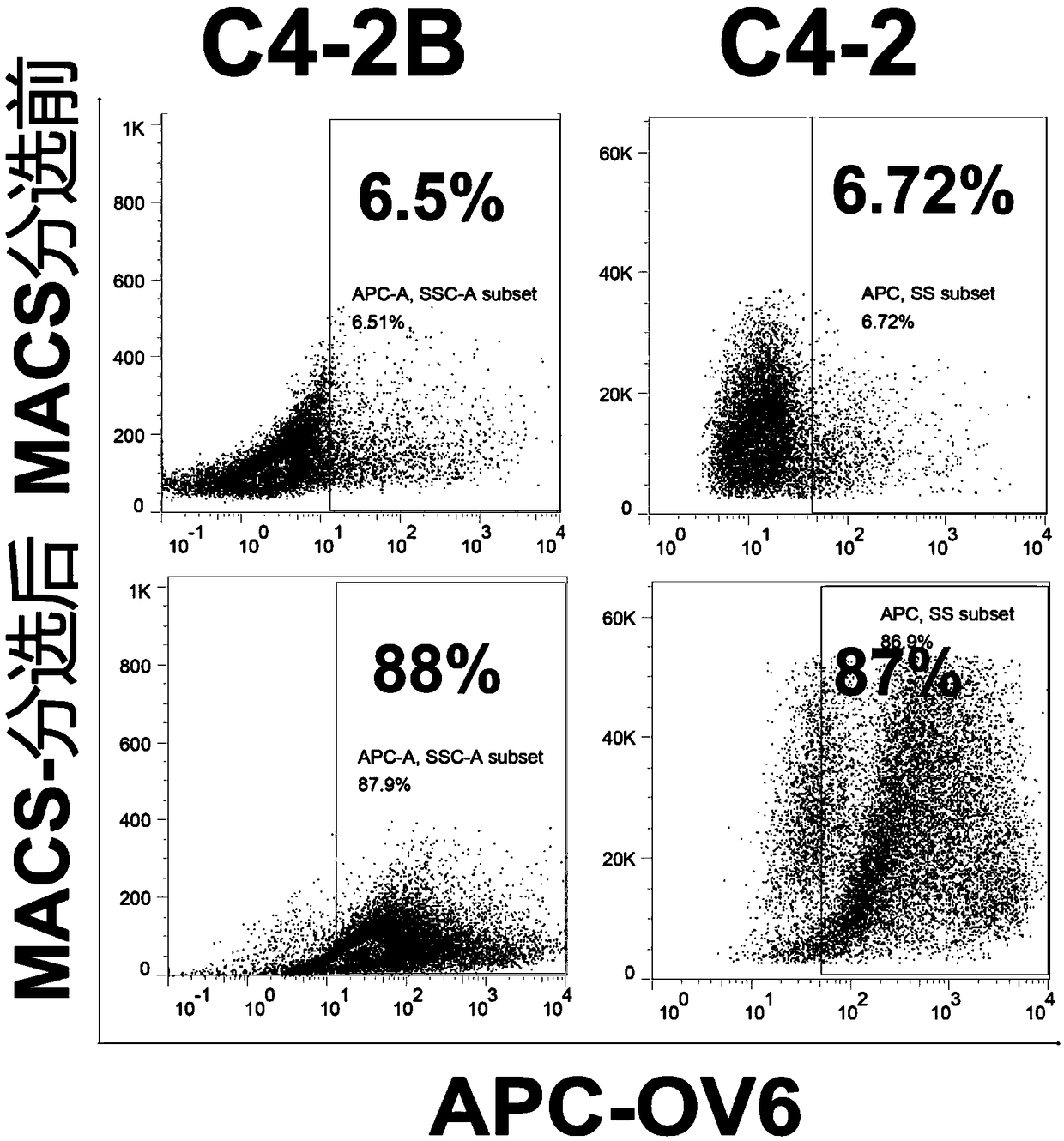
[0041] ② Magnetic-activated cell sorting (MACS) was performed using MiniMACS TM Cell Sorter (Miltenyi Biotec, Bergisch Gladbach, Germany), specifically labeled with mouse-derived OV6 antibody (1:50, MAB2020, R&DSystems, Minneapolis, MN, USA) C4-2 and C4-2B cells treated with ENZ for 30 minutes;
[0042] ③ Incubate C4-2 and C4-2B cells labeled with OV6 primary anti...
Embodiment 2
[0046] OV6 positive prostate cancer cell identification method
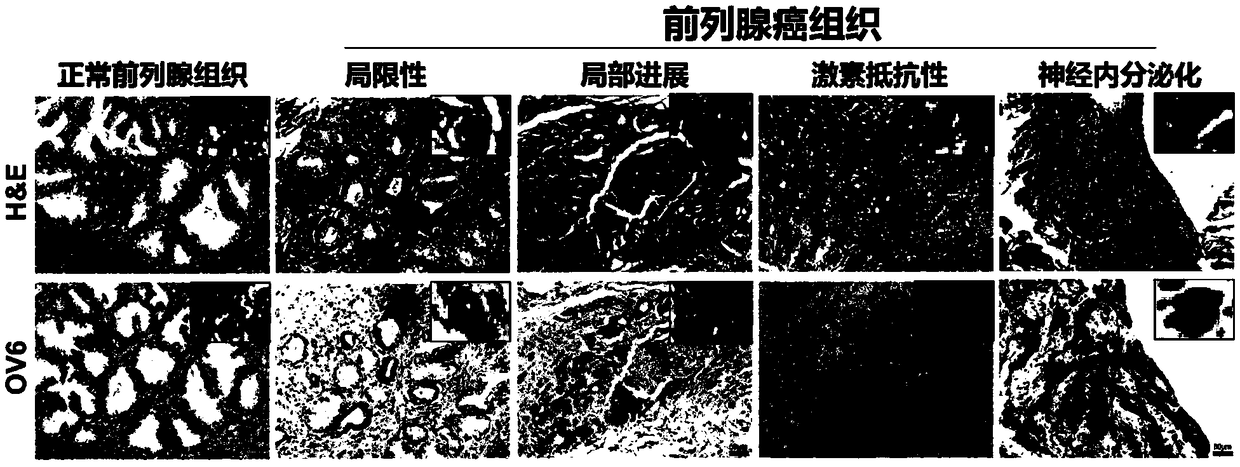
[0047] 1. Tissue sample collection and immunohistochemistry
[0048] Between 2012 and 2013, cohort 1: 78 patients with pathologically diagnosed prostate cancer at Changhai Hospital (Shanghai, China) and cohort 2: 67 patients with pathologically diagnosed prostate cancer at Changzheng Hospital (Shanghai, China) were collected , including clinical data, tissue samples and follow-up data.
[0049] The clinical data of prostate cancer patients included age at diagnosis, preoperative PSA (maximum value within 6 months before operation), postoperative pathological status including Gleason score (GS), pathological T stage, pathological N stage, and extracapsular invasion ( CP) Surgical margins (SM), peripheral nerve invasion (PNI) and seminal vesicle invasion (SVI). Time to biochemical recurrence (BCR) (standard: PSA ≥ 0.2 μg / μl), MRI, CT or ECT to determine disease recurrence, clinical observation endpoints include b...
Embodiment 3
[0060] Detection of Stem Cell Characteristics of OV6 Positive Prostate Cancer Cell Lines
[0061] 1. Detection of expression levels of stemness-related genes by real-time polymerase chain reaction
[0062] Total RNA was extracted from C4-2B, C4-2, sorted monocytes or THP-1 using RNAiso Plus (9109, Takara, Japan) according to the manufacturer's protocol. Reverse transcription was performed using PrimeScript One Step RT kit (RR037B, Takara, Japan) in the presence of random primers and reverse transcriptase. Amplification of the generated cDNA was carried out in SYBR GreenRealtime PCR Master Mix (QPK201, Toyobo, Japan) and ABI PRISM 7300HT Sequence Detection System.
[0063] Each measurement was performed in triplicate and the results were normalized by the expression of the β-actin gene. The fold change from the mean by 2 -△△Ct OK, the result is as follows Figure 8 shown, data are expressed relative to the OV6 - Fold change in C4-2B cells, indicating high expression of ste...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com