Method for synthesizing tagatose from cyanobacteria
A technology of tagatose and cyanobacteria, applied in the field of synthetic biology, can solve the problems of difficult industrialized large-scale production, many by-products under alkaline conditions, and difficult product separation, and achieve easy industrial transformation, good purity, scale and The effect of a small amount
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
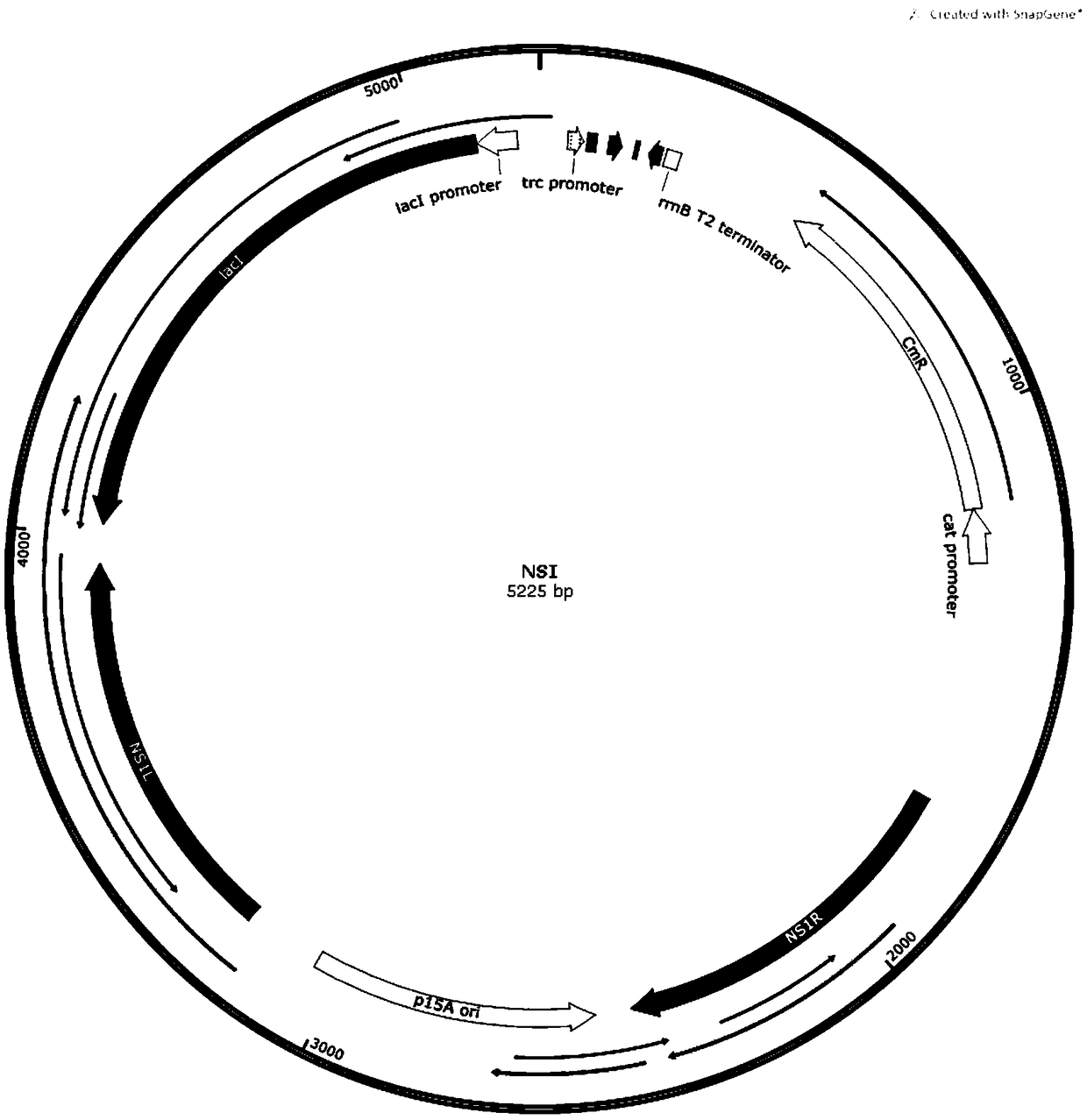
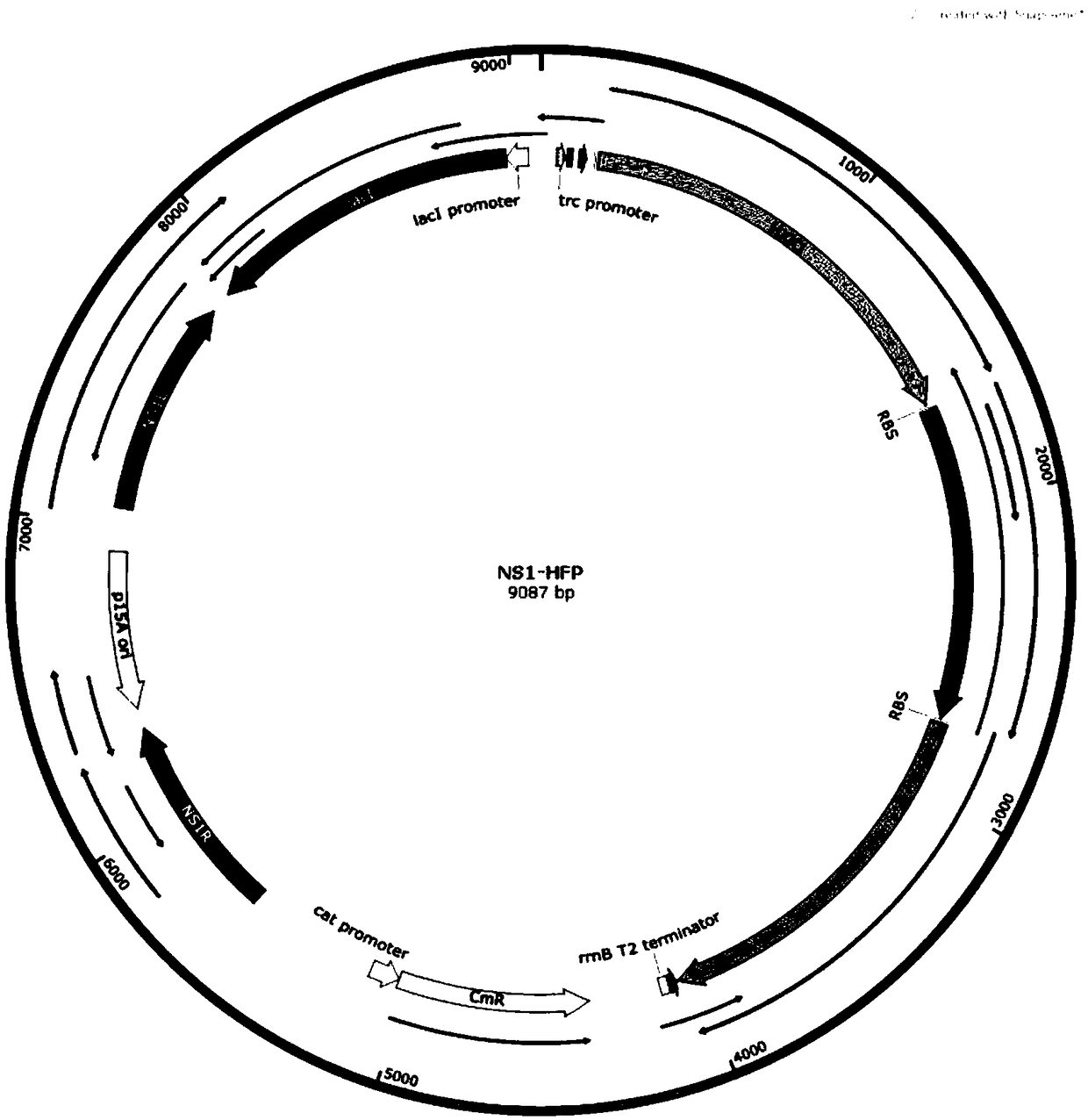
[0052] Plasmid construction process: According to the NSI gene sequence of cyanobacterium Syn7942, according to the attached figure 2 The plasmid map in the construction of the integration vector NSI. Then, according to the amino acid sequences of the genes HXK, FbaA and phytase provided in the present invention, the nucleic acid sequences of the corresponding genes are designed according to the codon preference of Cyanobacteria Syn7942 cells, and the genes HXK, FbaA and phytase are sent to commercial companies for synthesis, and the same size of about 20bp is designed. source arm, follow the attached image 3 In the plasmid map, the three genes were constructed on the integration vector NSI using a one-step cloning kit.
Embodiment 2
[0054] Conversion process:
[0055] 1) Take 2 mL of wild-type Syn7942 with a growth density OD730 of 0.8-1.2 in a centrifuge tube, then centrifuge at 10,000 rpm for 2 min, and remove the supernatant;
[0056] 2) Mix the precipitate in the previous step with 1mL 10mM sodium chloride solution, then centrifuge at 10000rpm for 2min, and remove the supernatant;
[0057] 3) Mix the precipitate in the previous step with 1 mL of sterilized anti-BG11-free liquid medium, then centrifuge at 10,000 rpm for 2 min, and remove the supernatant;
[0058] 4) Mix the precipitate in the previous step with 100 uL of sterilized anti-BG11-free liquid medium, and then add 200 ng of plasmid DNA to it. Thoroughly seal the centrifuge tube with tinfoil (protected from light), then incubate in a shaker at 100 rpm at 30°C for 10 hours;
[0059] 5) Apply all the cyanobacteria in the centrifuge tube in the previous step to the BG11 solid medium containing 25ug / mL chloramphenicol, put the plate in a light i...
Embodiment 3
[0062] Verification of transgenic cyanobacteria and product detection process:
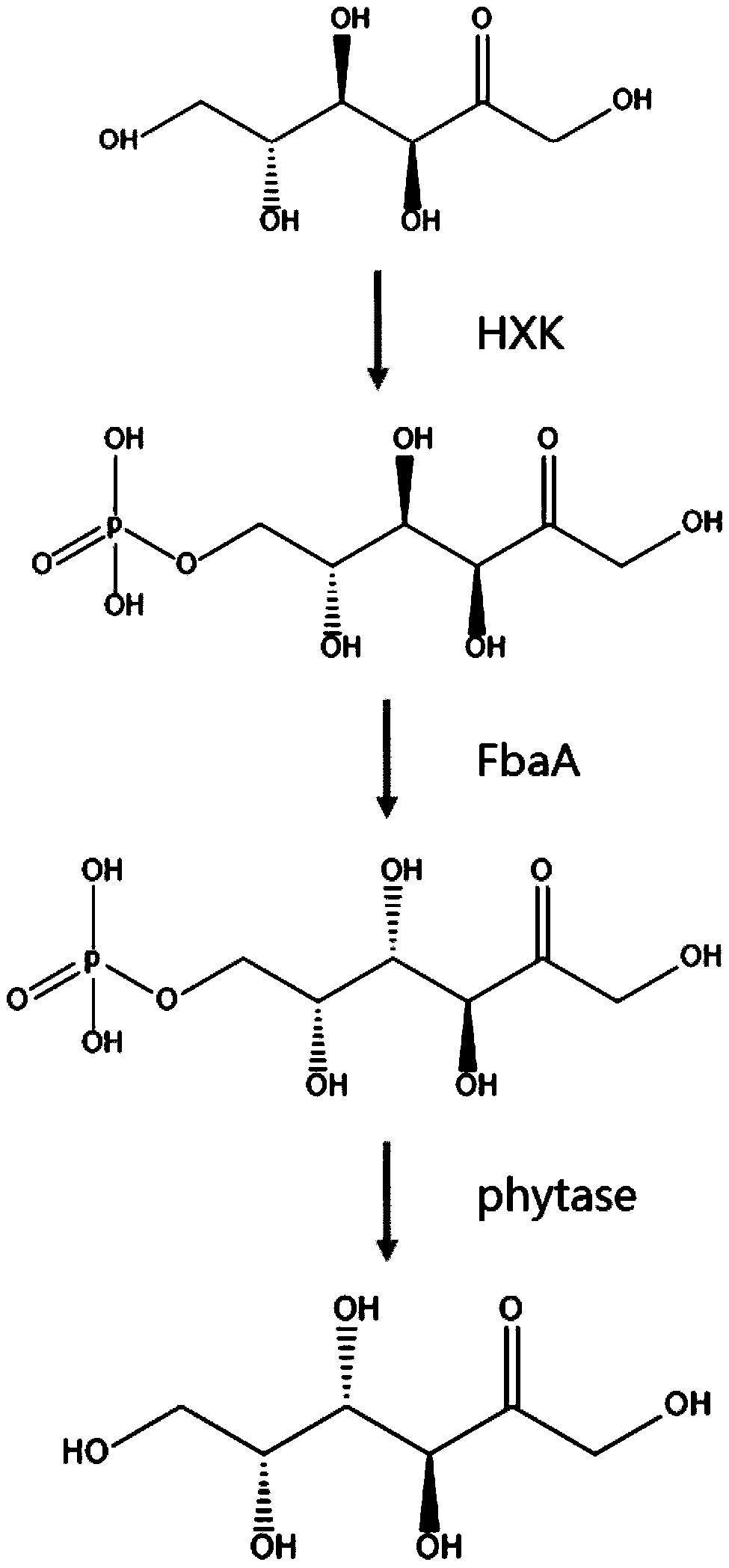
[0063] Pick about 10 monoclonal cyanobacterial plaques grown on BG11 solid medium containing 25ug / mL chloramphenicol, insert them into 5mL25ug / mL chloramphenicol BG11 liquid medium for culture, and wait until the cyanobacteria are cultivated until the OD730 is 0.5 , extract the genome, verify the three target genes of HXK, FbaA and phytase by PCR with the designed primers, and send the products to sequence by PCR to determine whether the gene sequence is correct; transfer the correct verified monoclonal transgenic cyanobacterium Syn7942 into 100mL BG11 Culture in liquid medium under light. When OD730 is S, add IPTG to induce the expression of three genes, HXK, FbaA and phytase. The expressed enzymes will synthesize tagatose with fructose produced by photosynthesis of cyanobacteria, and then take the induced The transgenic cyanobacteria are cultured for 1 day, 3 days, and 5 days, and the cyanobacte...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com