Method for constructing expression library of cell penetrating peptide by utilizing bacteriophage display technology
A phage display and penetrating peptide technology, applied in the field of biomedicine, can solve the problems of low positive rate, labor and time consumption, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0068] A method for constructing a cell-penetrating peptide expression library using phage display technology, using phage display technology, combined with the whole-cell screening mode, to screen polypeptides with cell membrane penetrating activity; and then using the screened cell-penetrating peptides to construct cell-penetrating peptides Prokaryotic expression library.
[0069] The principle of the phage display technology described in the present invention is to recombine the DNA fragment encoding the polypeptide with the gene encoding the phage capsid protein, and express it on the surface of the phage in the form of fusion protein. Based on the affinity between biomolecules and target molecules, through the repeated process of adsorption-elution-amplification, phages containing specific binding to target molecules are screened from phage libraries expressing various foreign proteins, and then enriched, Amplification and gene sequence determination to obtain the amino a...
Embodiment 2
[0108] Example 2: Screening of cell-penetrating peptides by phage display technology
[0109] Titer determination of the first step phage display cyclic heptapeptide library
[0110] Inoculate a single colony of E.coli (Escherichia coli) ER2738 in 5ml LB medium, shake vigorously (225rpm) at 37°C, and cultivate to mid-logarithmic phase (OD 600 value around 0.5). Thaw the top agar in a microwave oven and store at 45°C until use. Divide into 3-4ml aliquots before use, pour into sterilized glass test tubes, one tube for each phage dilution. Take 1 μl of phage random cyclic heptapeptide library, and make 10-fold serial dilution in LB, the dilution range is 10 9 -10 11 (The dilution range of each round in the screening process is: the amplified phage culture supernatant 10 9 -10 11 ; unamplified panning isolate 10 2 -10 4 ). When the bacterial culture reached mid-log phase, divide into 200 μl aliquots in microcentrifuge tubes, one for each phage dilution. Add 10 μl of diff...
Embodiment 3
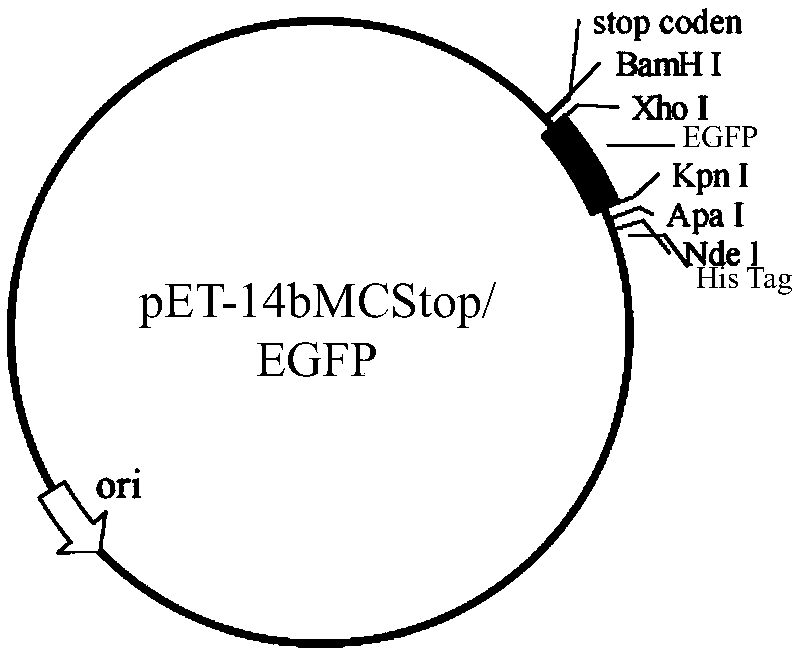
[0118] Example 3: Construction of a cell-penetrating peptide prokaryotic expression library
[0119] The polypeptide DNA sequence in the prokaryotic expression library of the present invention is derived from the total DNA of a random cyclic heptapeptide library screened by phage display technology. By comparing the recovery rate and enrichment factor of phage random heptapeptide library by four rounds of whole-cell screening, we found that the enrichment factor reached the maximum after the third round of screening. The decline of the enrichment factor in the fourth round suggests that the screening has approached saturation, and continued screening may lead to a decrease in diversity and loss of data. Therefore, the phage DNA extracted from the third round of screening was selected to construct the prokaryotic expression library of Hela cell-penetrating peptides. This step is a key control point that needs to be considered emphatically in the establishment of this method. ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| electrical resistance | aaaaa | aaaaa |
| strength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com