Method for identifying Shida tea varieties through SSR fingerprint spectrum
A technology of fingerprinting and persimmon tea, applied in the field of molecular biology, can solve problems such as difficult to distinguish, and achieve high accuracy, high polymorphism, and good stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
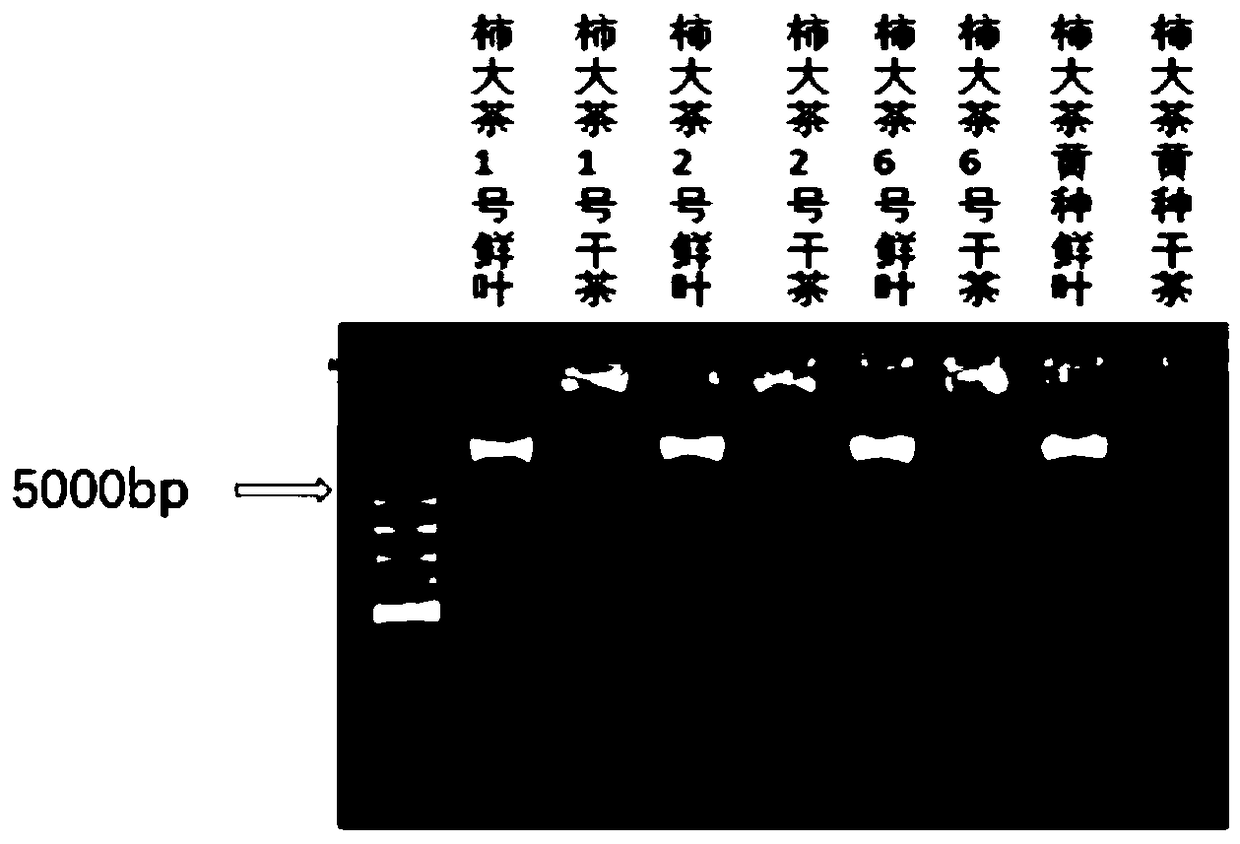
[0028] The extraction of embodiment 1 tea fresh leaf total DNA
[0029] (1) The selected tea tree varieties: No. 1 Shidacha; No. 2 Shidacha; No. 6 Shidacha; Huangzhong Shidacha. Provided by the Industry-University-Research Base of Guohe Modern Agriculture Demonstration Zone of Anhui Agricultural University
[0030] (2) Using the selected tea tree species as material, the CTAB method was used to extract the total DNA, and the specific operation process was:
[0031] ① Take a 2ml centrifuge tube, add 0.1g of ground dry tea powder, add 900mL of CTAB extract (preheated to 65°C in advance), add 20mL of β-mercaptoethanol, bathe in water at 65°C for 20min, shake gently up and down several times every 10min , then add 10mL RNAase and place in a 65°C water bath for 15min, shake up and down several times every 5min.
[0032] ② Centrifuge at 13000r / min for 10min, take 800mL of supernatant, add 400mL Tris to balance phenol, add an equal volume (800mL) of chloroform: isoamyl alcohol to 2...
Embodiment 2
[0036] The selection of embodiment 2SSR site and the preliminary screening of primer
[0037] Among the 100,000 sequences containing SSR sites, the SSR sites with hexanucleotides as repeating units were more and more polymorphic. Therefore, 4,000 sequences containing SSR sites were selected according to the following principles, and 450 pairs of primers (synthesized by General Biology) were successfully designed. The principles of SSR primer screening are as follows:
[0038] ①The length of the primer is 18-22bp, and the target fragment is between 250bp-380bp.
[0039] ②The GC content is 40%-60%, avoid three or four consecutive bases in the primer sequence;
[0040] ③The annealing temperature should be between 45°C and 55°C, preferably around 50°C, and the difference between the Tm values of the upstream and downstream primers should not be greater than 4°C;
[0041] ④ Avoid more than 3 consecutive bases at the 3' end of the primer, and try to avoid primer dimers and hairp...
Embodiment 3
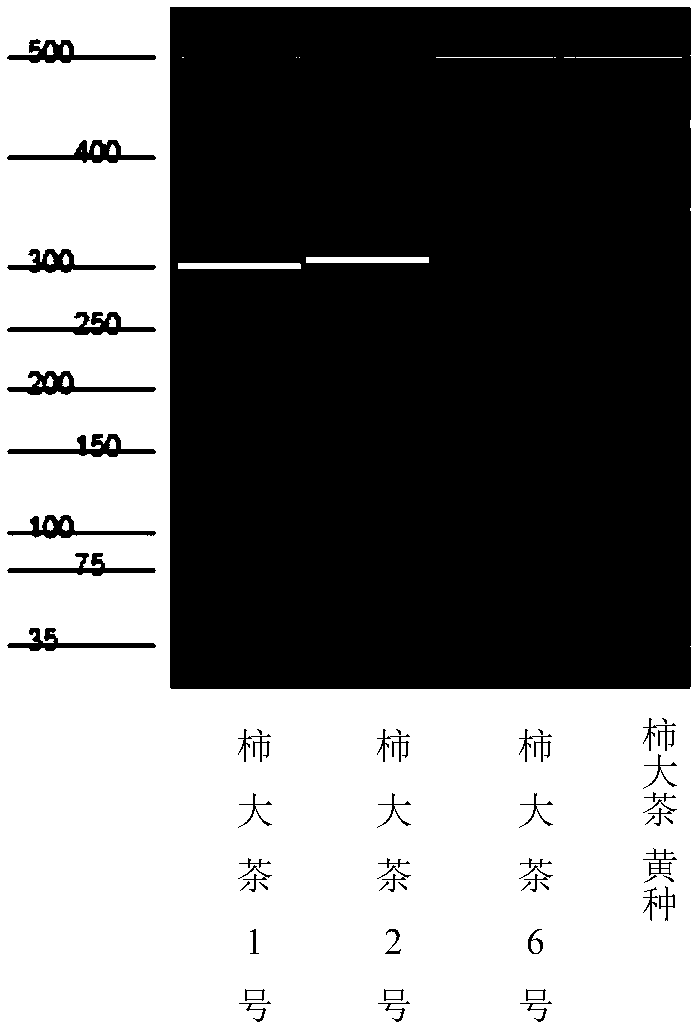
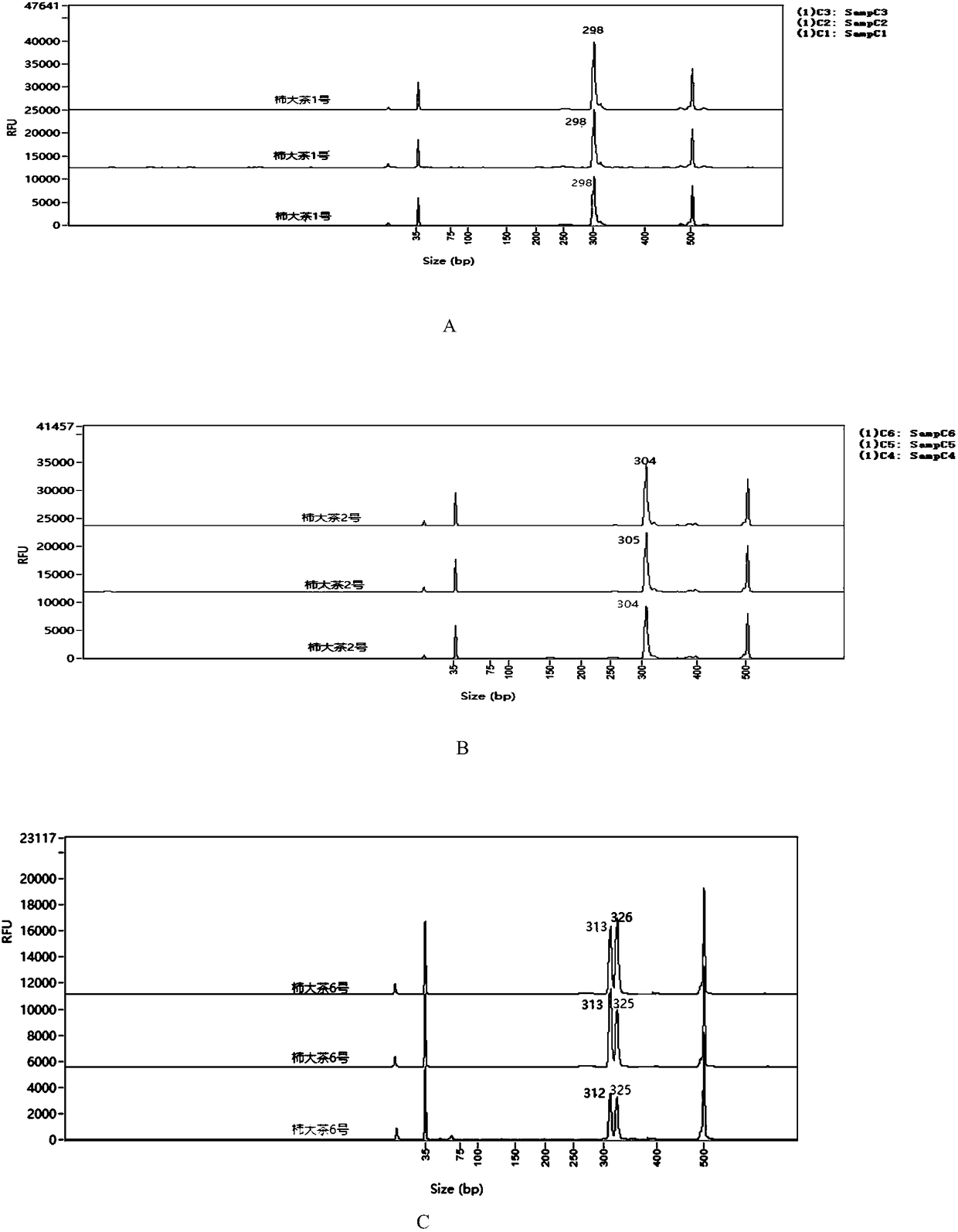
[0042] Embodiment 3PCR amplification and Fragment Analyzer TM Automatic capillary electrophoresis system primary screening
[0043]The total reaction system is 20μL, including 2μL of 50ng / μL DNA solution, 0.5μL of 10μM upstream and downstream primers, 2μL of 10×EasyTaq Buffer, 1units of EasyTaq DNA Polymerase, 2μL of 2.5mM dNTP, ddH 2 O 12.8 μL. (Reagents are from Transgeng Biological Company)
[0044] The PCR reaction program was: pre-denaturation at 94°C for 5 min, 30 cycles (denaturation at 94°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 30 s), extension at 72°C for 10 min, and storage at 4°C. The annealing temperature was determined according to each primer. PCR amplification and capillary electrophoresis primary screening. Specifically, 2 microliters of the PCR product was taken and passed through the Fragment AnalyzerTM automatic capillary electrophoresis system for primary screening, and the primary screening of primers was performed by comparing with...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com