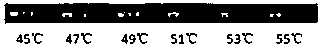Specific primer for identifying poecilobdella manillensis based on molecular biological method
A molecular biology and specific technology, applied in the field of molecular biology, can solve the problems of difficult and time-consuming identification of Hirudo phenanthae medicinal materials, and achieve the effect of reducing the time of comparative analysis, reducing operation time, and reducing professional restrictions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] A pair of specific primers based on specific amplification of molecular biology to identify Hirudo phenanthrae, the upstream primer is: 5'-TCTGGCTCTTAGGCTTCG-3', the downstream primer is: 5'-GTTGTCTTCCTGGCTGTT-3', provided by Shanghai Jierui Bioengineering Co., Ltd. Company Synthesis.
[0028] Using the above primers to quickly identify Hirudo phenanthae, the samples to be tested were selected from the leech samples numbered SZ-1~SZ-20 in Table 1, and parallel experiments were carried out to identify whether the leech samples numbered SZ-1~SZ-20 were Hirudo phenatus. ;Specific steps are as follows:
[0029] (1) Extraction of total DNA: Take 20-30 mg of the sample to be tested, grind the dry product with MM400 ball mill, cut the fresh sample with surgical scissors, and use the DNeasy Blood & Tissue Kit genome extraction kit from QIAGEN Company to perform DNA extraction;
[0030] (2) Polymerase chain reaction: 20 μL of PCR reaction system includes 10 μL of 2×Taq Master M...
Embodiment 2
[0035] The annealing temperature of the specific primers in Example 1 was investigated. A total of 6 annealing temperatures of 45°C, 47°C, 49°C, 51°C, 53°C, and 55°C were investigated. According to the investigation results, 49°C was determined to be The optimal annealing temperature of primer 7 to specifically amplify the sequence of B. phenanthrene. Specific steps are as follows:
[0036] (1) Extraction of total DNA: randomly select three batches of samples numbered SZ-10, SZ-14, and SZ-18, take 20-30 mg of the sample to be tested, grind the dry product with MM400 ball mill, and cut the fresh sample with surgical scissors , using QIAGEN's DNeasy Blood&Tissue Kit genome extraction kit for DNA extraction;
[0037] (2) Polymerase chain reaction: 20 μL of PCR reaction system includes 10 μL of 2×Taq Master Mix buffer, 0.4 μL of 25 μmol / L upstream and downstream primers, 1 μL of 50 ng / μL DNA template, and 8.2 μL of sterilized double-distilled water. The PCR reaction conditions a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com