Kit and method for extracting collybia radicata genome DNA (Deoxyribonucleic Acid)
A genome and kit technology, applied in the field of molecular biology, can solve the problems of short DNA fragments, toxic reagents, long time-consuming, etc., and achieve the effects of good integrity, increased binding rate, and shortened extraction time.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
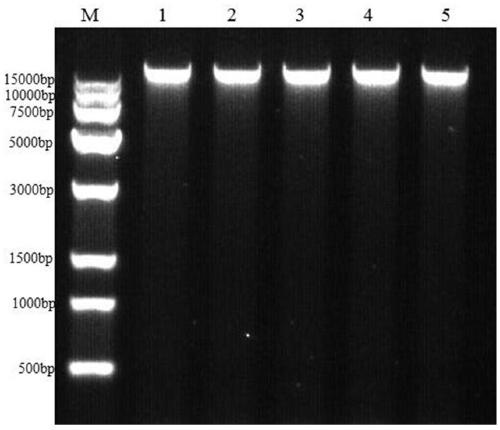
[0034] Embodiment 1, adopt the kit of the present invention to extract the genomic DNA of rhizome rhizome mycelium and fruiting body
[0035] The composition of the kit includes:
[0036] (1) Balance Buffer, which contains the following components: 1% Na 2 SiO 3 9H 2 O, 1% NaOH;
[0037] (2) Lysis Buffer, which contains the following components: 5.0M guanidine hydrochloride, 25% isopropanol and 1% PVP K30;
[0038](3) Wash Buffer, containing the following components: 20 mM NaCl, 20 mM Tris-HCl, 85% ethanol, pH 7.5;
[0039] (4) Elution Buffer, containing the following components: 10 mM Tris-HCl, pH 8.5;
[0040] (5) DNA adsorption column.
[0041] Further, the DNA adsorption column is an aluminum silicate ceramic fiber membrane DNA adsorption column.
[0042] Collect respectively the mycelium and the fruiting body of Rhizoma Rhizome under the same culture condition, and concrete steps are as follows:
[0043] (1) Add 200 μL of Balance Buffer to the aluminum silicate ce...
Embodiment 2
[0050] Example 2 Using the kit and method of the present invention to extract genomic DNA from edible fungi
[0051] Kit composition includes:
[0052] (1) Balance Buffer, which contains the following components: 1% Na 2 SiO 3 9H 2 O, 1% NaOH;
[0053] (2) Lysis Buffer, which contains the following components: 5.0M guanidine hydrochloride, 25% isopropanol and 1% PVP K30;
[0054] (3) Wash Buffer, containing the following components: 20 mM NaCl, 20 mM Tris-HCl, 85% ethanol, pH 7.5;
[0055] (4) Elution Buffer, containing the following components: 10 mM Tris-HCl, pH 8.5;
[0056] (5) DNA adsorption column.
[0057] Further, the DNA adsorption column is an aluminum silicate ceramic fiber membrane DNA adsorption column.
[0058] Collect the fruiting body, mycelium, and Agaricus blazei, Cordyceps militaris, and Grifola frondosa fruiting body samples respectively, and use the kit and method of the present invention to extract the genomic DNA of these samples. The specific ste...
Embodiment 3
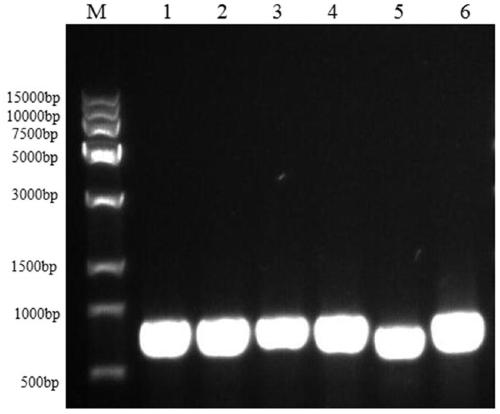
[0065] Embodiment 3, long root mushroom ITS sequence PCR amplification
[0066] Using general primers ITS-4 / 5 for the ITS fragment of edible fungus, the genomic DNA of the extracted fruiting bodies, mycelia, and dry samples of Rhizoma lanceolata, as well as the fruiting bodies of Agaricus blazei, Cordyceps militaris, and Grifola frondosa were amplified by PCR.
[0067] The primer sequences are:
[0068] ITS-4: 5’-TCCTCCGCTTATTGATATGC-3’
[0069] ITS-5: 5’- GGAAGTAAAAGTCGTAACAAGG-3’
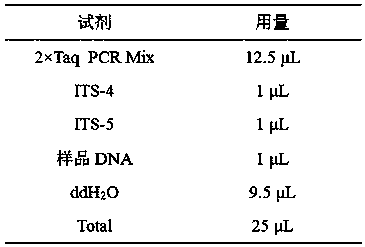
[0070] The reagents used for PCR amplification were purchased from Beijing Quanshijin Biotechnology Co., Ltd., and the reaction system was:
[0071]
[0072] The conditions of the PCR reaction are: 94°C 7min → [94°C 30s → 56°C 30s → 72°C 30s] × 34cycles → 72°C 10min → 4°C.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com