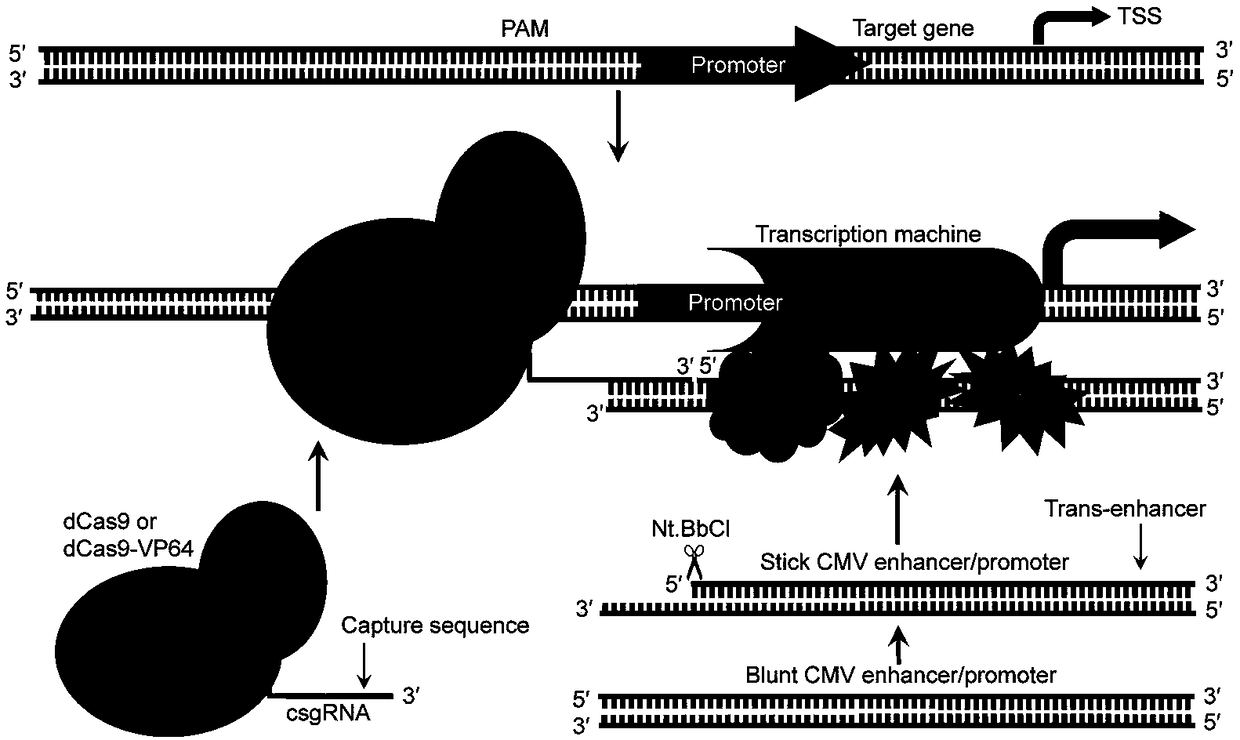
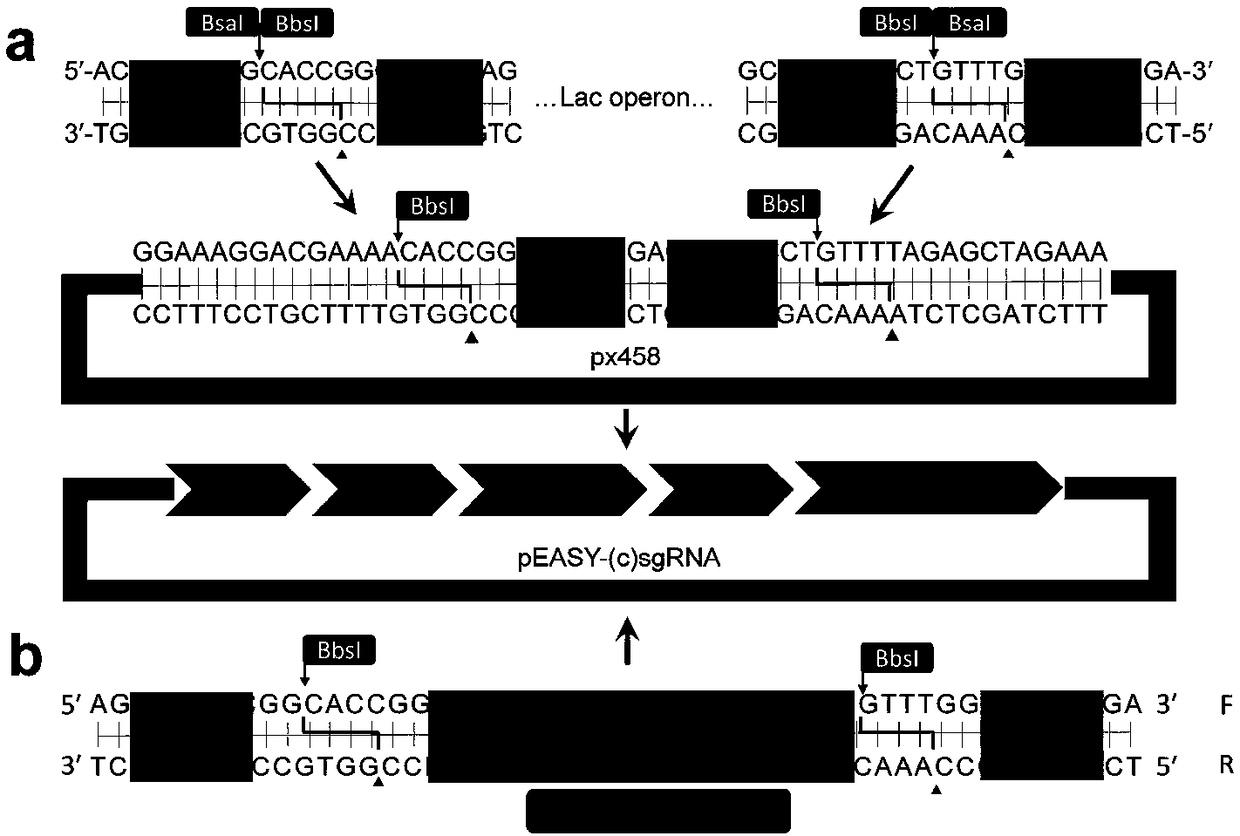
Method for activating gene expression by CRISPR-assisted trans-enhancer and application thereof
A gene expression and target gene technology, applied in the field of biomedicine, can solve problems such as low-efficiency gene activation activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] vector construction
[0059] experimental method:
[0060] To apply the CRISPR / dCas9 expression system for transfection in cells, a plasmid containing sgRNA driven by a U6 promoter was constructed. Using Pfu high-fidelity polymerase (Transgen, AS221-01), using primers Lac-px-F (Table 1) and Lac-px-R (Table 1) cloned from pEASY-Blunt-simple (Transgen, CB101-01) The lac operator sequence with BbsI and BsaI sites at both ends. This lac operator sequence was ligated into px458 (Addgene plasmid ID: 42230) to construct px458-lac. The ligation product was transformed into competent cells DH5α, and then blue colonies were screened. Verify px458-lac by sequencing. Three flanking sequences were then designed. Using the forward primer (U6-F; Table 1) and one of three reverse primers U6-1-R (Table 1), U6-2-R (Table 1), and U6-3- R (Table 1), flanking sequences were added to the 3'-end of the gRNA scaffold sequence by PCR amplification, where the gRNA scaffold sequence cloned ...
Embodiment 2
[0108] Effect of capture sequence on sgRNA function
[0109] experimental method:
[0110] DNA cleavage with Cas9 / csgRNA: Select sgRNA targeting the HNF4α promoter sequence. sgRNAs were prepared by in vitro transcription using T7 RNA polymerase (M0251S, NEB). The sgRNA transcription template was prepared by PCR amplification of the sgRNA coding sequence cloned in the sgRNA expression plasmid (pEASY-csgRNA) with the forward primer HNF4α-T7-F (Table 5); the forward primer contained the T7 promoter sequence (TAATACGACTCACTATAG, Transcription starts at 3'G), and one of four reverse primers U6-R, U6-1-R, U6-2-R and U6-3-R (Table 1). A normal sgRNA (HNF4α-sgRNA) and three csgRNAs (HNF4α-csgRNA) were prepared. A 732-bp HNF4α promoter fragment was amplified from pEZX-HP-ZsGreen-A by PCR using primers HNF4α-sP-F and HNF4α-sP-R (Table 5). The Cas9 digestion reaction system (30 μL) consists of 1×Cas9 nuclease reaction buffer, 1 μM Cas9 nuclease (NEB, M0386T) and 300 nM HNF4α-sgRNA or...
Embodiment 3
[0116] Activation of exogenous reporter genes through trans-enhancers
[0117] experimental method:
[0118] Cell culture and transfection:
[0119] 293T, HepG2, A549, SKOV3, HT29, PANC-1, and HeLa were obtained from the Cell Resource Center, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, and incubated at 37°C and 5% (v / v) CO 2 incubator culture. Cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) or Roswell Park Memorial Institute (RPMI) 1640 medium containing 10% FBS, 100 U / mL penicillin and 100 μg / mL streptomycin. When cells are confluent >70% in each well of a 12-well plate, use 800ng total DNA, including 500ng plasmid (pcDNA-dCas9, pcDNA-dCas9-VP64 or pcDNA-dCas9-VPR), 150ng linear sgRNA expression template, ( U6-sgRNA or U6-csgRNA) and 150ng linear CMV, dissolved in Lipofectamine 2000 (ThermoFisher Scientific) liposomes were transfected using the following protocol. For each transfection, when cells grow to 4 x 10 5 At a density...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com