Detecting method for captopril through light emitting silver clusters synthesized by G insertion sequences
A technology of fluorescence detection and sequence, which is applied in the directions of measurement devices, fluorescence/phosphorescence, material excitation analysis, etc., can solve the problems of low sensitivity, high cost, environmental pollution, etc., and achieve the effect of high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Prepare the DNA-AgNCs solution using the following conditions:
[0031] (1) Preparation of DNA-AgNCs solution: Take 20 μL of phosphate buffer, add DNA aqueous solution with a final concentration of 5 μM, and then add 2.0 μL of AgNO with a concentration of 3.0 mM 3 Aqueous solution, stirred for 30s, kept at 4°C for 20min, then added 2.0μL fresh NaBH 4 aqueous solution and vigorously stirred for 1 min, and the mixture was stored at room temperature in the dark for 9 h to prepare a DNA-AgNCs nanocluster solution;
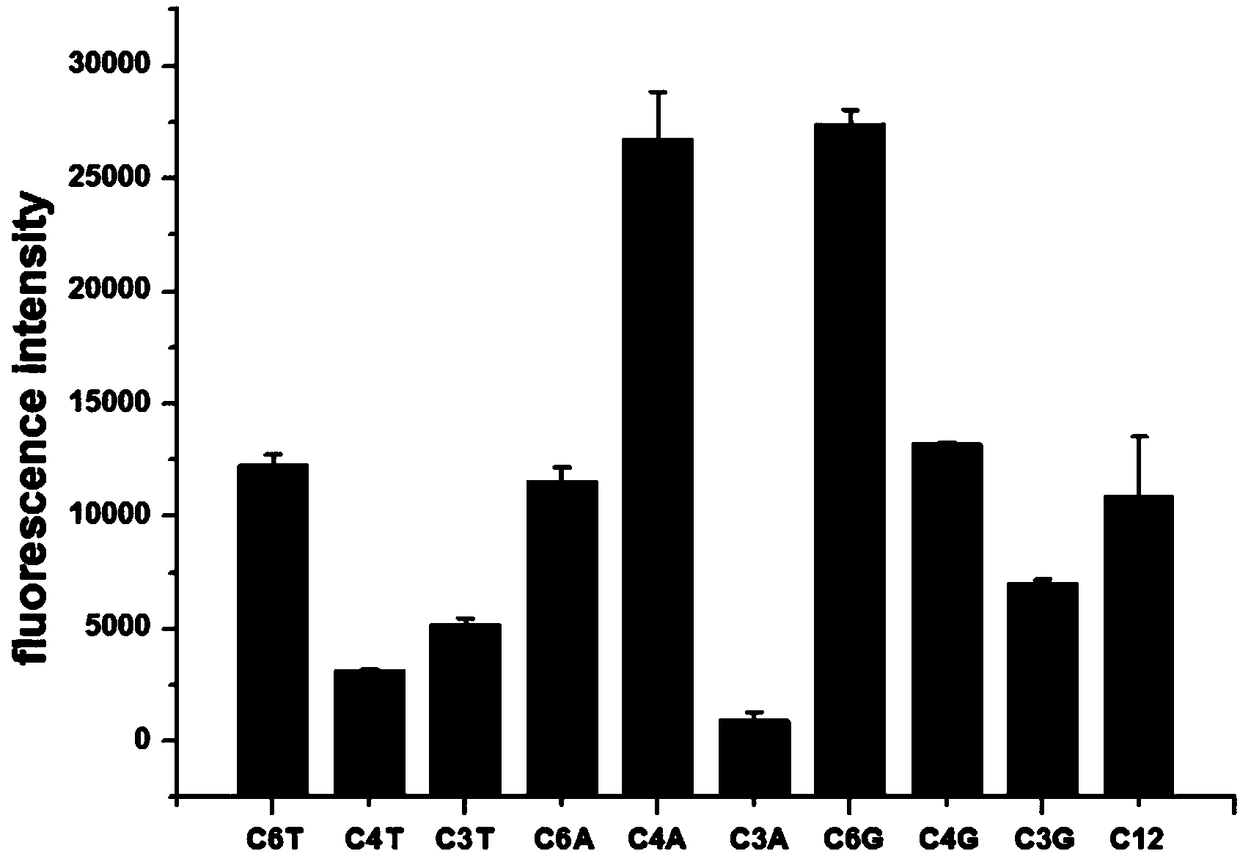
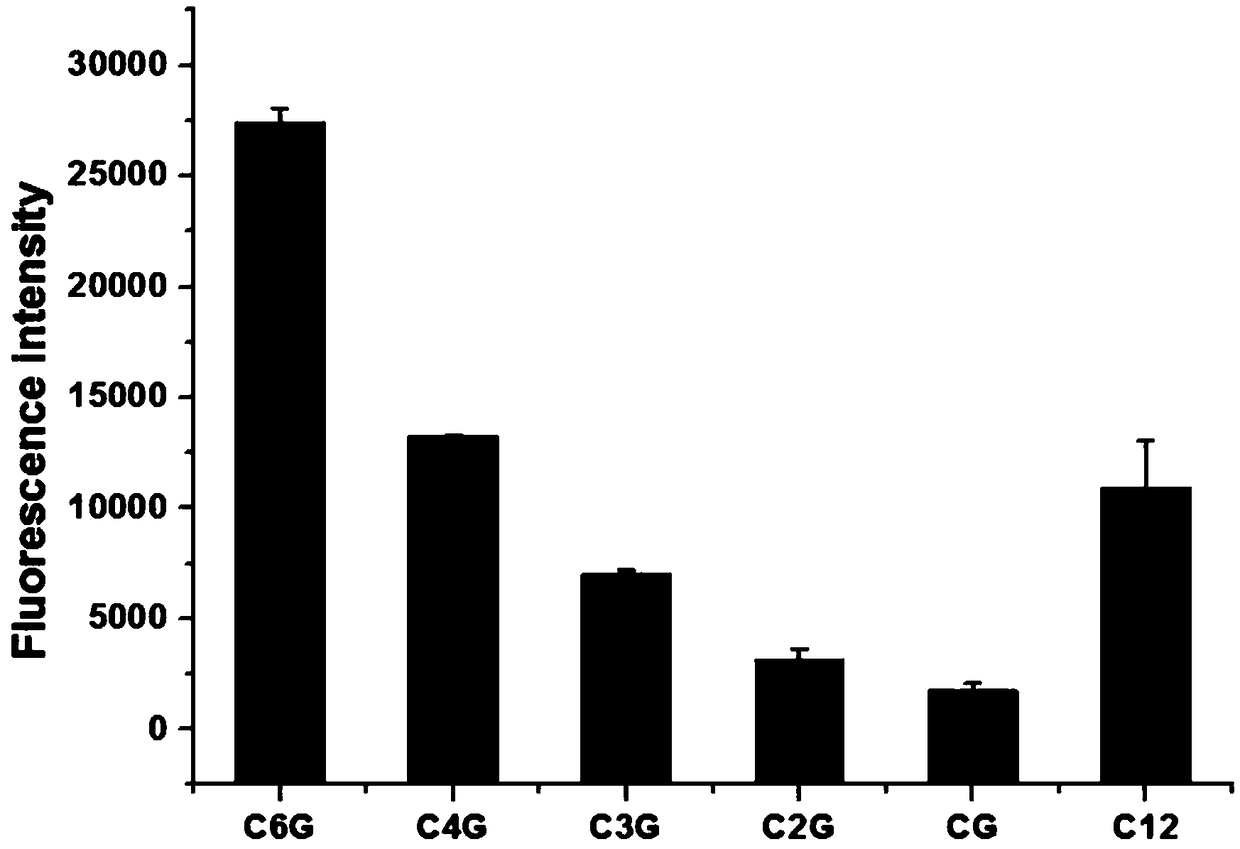
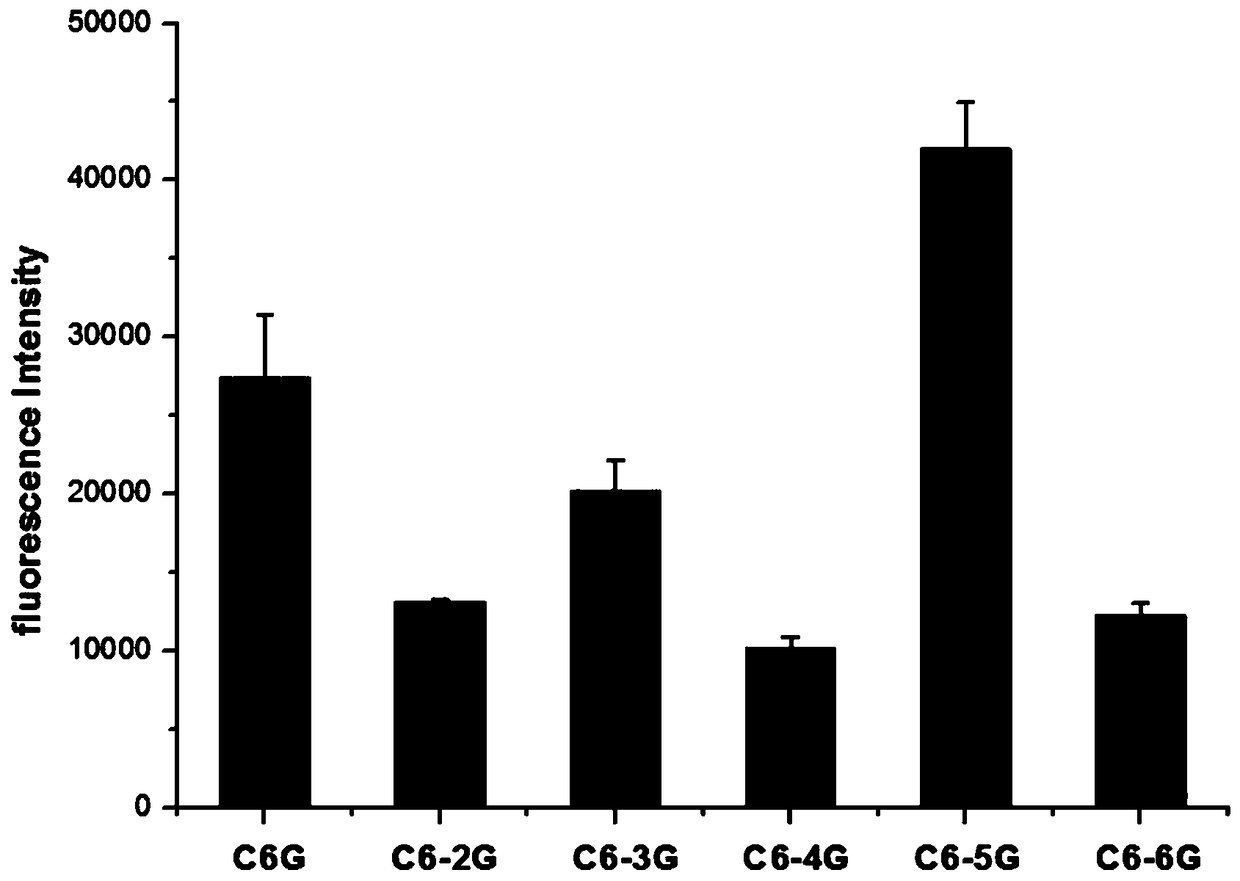
[0032] (2) Screening of DNA sequences: select different C-rich base DNA sequences in Table 1, synthesize DNA-AgNCs according to step (1), measure its fluorescence intensity at the maximum emission wavelength (670nm), the results are as follows figure 1 As shown, the fluorescence intensities corresponding to the C4A and C6G sequences are relatively strong, 26730 and 27394 respectively. It can be seen that the C6G sequence has the highest intensity, so the G inse...
Embodiment 2
[0045] Add different concentrations of captopril to the 2.5 μM C6-5G-AgNCs nanocluster solution prepared in Example 1, the total reaction volume is 400 μL, and the final concentrations of captopril are 0, 0.05, 0.1, 0.2 . Figure 6 As shown, there is a good linear relationship in the captopril concentration range (0~1μg / mL), and the linear equation is y=-42774.68x+45953.36(R 2 = 0.9446). The detection sensitivity can reach 0.05μg / mL, and the detection linear range is 0.05μg / mL~5μg / mL.
Embodiment 3
[0047] The total reaction volume was 400 μL, and the concentration of C6-5G-AgNCs was 2.5 μM. Captopril at a concentration of 1 μg / mL, starch at 50 μg / mL, and Dextrin, 10 μg / mL maltose, 20 μg / mL gelatin, 10 μg / mL sucrose, 50 μg / mL lactose, 10 μg / mL glucose, and as a blank control, after fully reacting for 20 minutes, measure at the emission wavelength of 670nm Fluorescence intensity to determine the selectivity of C6G5-AgNCs to captopril detection, the results are as follows Figure 7 As shown, only the C6-5G-AgNCs added with captopril was significantly quenched in the fluorescence. The fluorescence intensity of the blank control group was 72251, that of the captopril group was 7737, that of the starch group was 63195, and that of the lactose group was 50311. and in Figure 7 In (b), it can be directly observed that the brightness of the sample added with captopril is much weaker than that of the other eight, indicating that the selectivity of C6G5-AgNCs for the detection of ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com