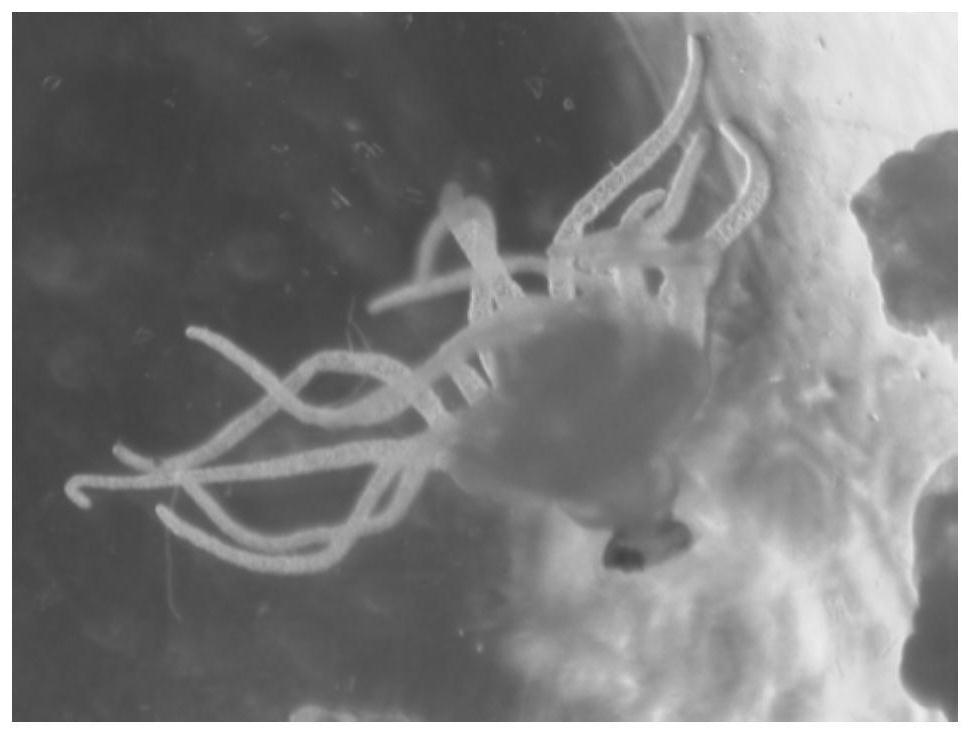
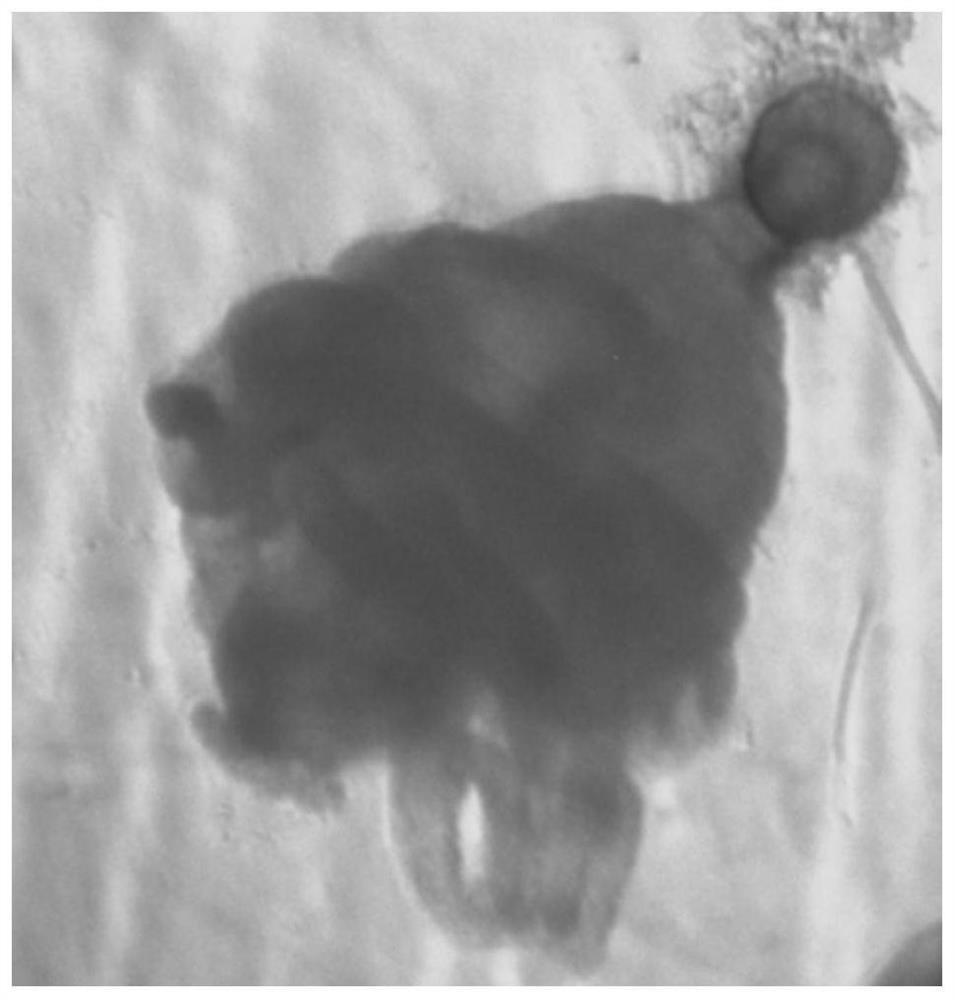
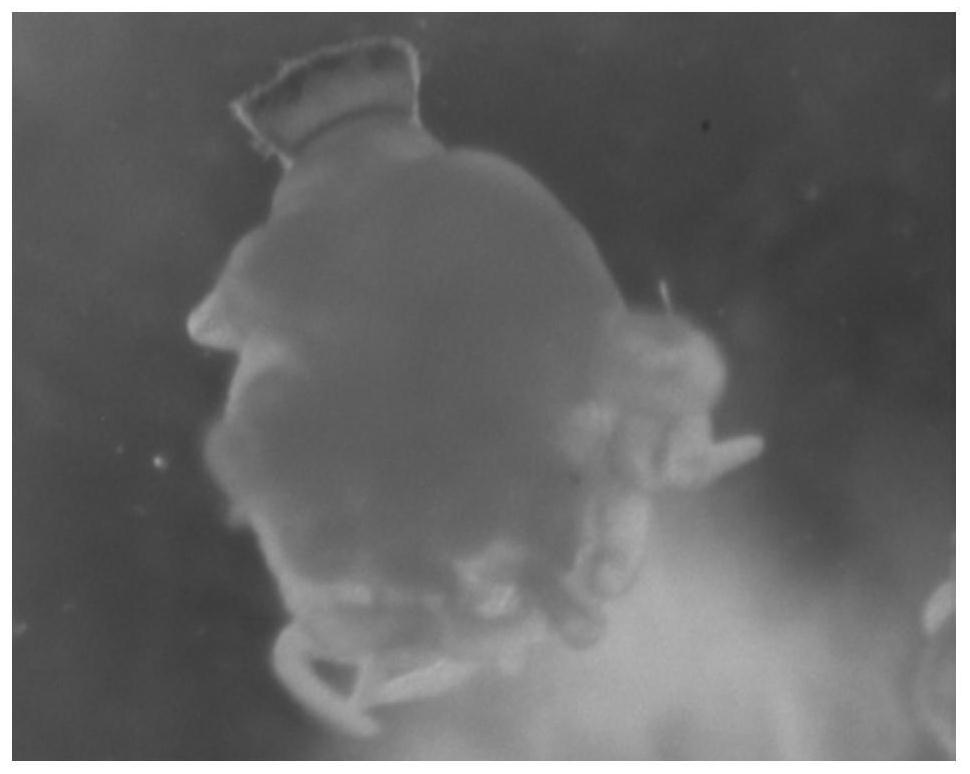
Whole-mount in situ hybridization detection method for marine jellyfish polyps
A detection method, in situ hybridization technology, applied in the field of cytogenetics and molecular biology, can solve the problems of overall in situ hybridization detection difficulties, weak probe specificity, complex microstructure, etc., to improve flexibility and operability Sexuality, individual integrity, and strong specificity of hybridization signals
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] Step A1, collection of jellyfish polyps
[0056] Take the corrugated board of the jellyfish polyp, put a piece of corrugated board into the filtered seawater, scrape off the polyp for observation, and inhale the polyp at the same developmental stage into the same centrifuge tube. First rinse the polyps with filtered seawater, then rinse with sterilized seawater, and absorb excess sterilized seawater. The polyps in the centrifuge tube were drawn into RNase-free collection tubes in stages, and labeled (date, time, developmental stage). The collection tubes containing the jellyfish polyps were centrifuged quickly and excess sterile seawater was aspirated.
[0057] Step A2, fixation of jellyfish polyps
[0058] Prepare the mixed solution with reference to Table 1 to obtain a clear solution.
[0059] Table 1. Mixed solution preparation table
[0060] Sterilized seawater PBS (0.1M, pH7.4) Mixture 1 Dilute to 100mL / Mixture 2 80mL Dilute to ...
Embodiment 2
[0086] Step B1: Hydration and digestion of jellyfish polyps
[0087] According to the method described in the above-mentioned Example 1, the jellyfish polyps that were fixed and dehydrated were obtained, and the polyps were transferred into a four-well cell culture plate for hydration, and passed through 70% ethanol aqueous solution, 50% ethanol aqueous solution, and 30% ethanol successively. Rinse with % ethanol NaPBS-Tween solution for 10 min each, and finally rinse with NaPBS-Tween three times, each time for 6 min. Digestion solution was added to each well and left to digest at 25°C for 8 min. To stop the digestion, add stop solution (100mg / mL glycine aqueous solution, the volume ratio of the above digestion solution is 1:50) to each well, quickly absorb it; then dilute 100g / mL glycine aqueous solution with glycine solution (2mg / mL, NaPBS-Tween) Rinse for 5min; finally rinse twice with NaPBS-Tween, 5min / time.
[0088] Step B2: Fixation and acetylation of jellyfish polyps ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com