Construction method of pediococcus acidilactici for producing L-lactic acid through co-fermentation of glucose and xylose
A technology for the construction of Pediococcus lactis, which is applied in the field of Pediococcus lactis construction, can solve the problems of no actual proof of engineering strains and increased fermentation costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment example 1
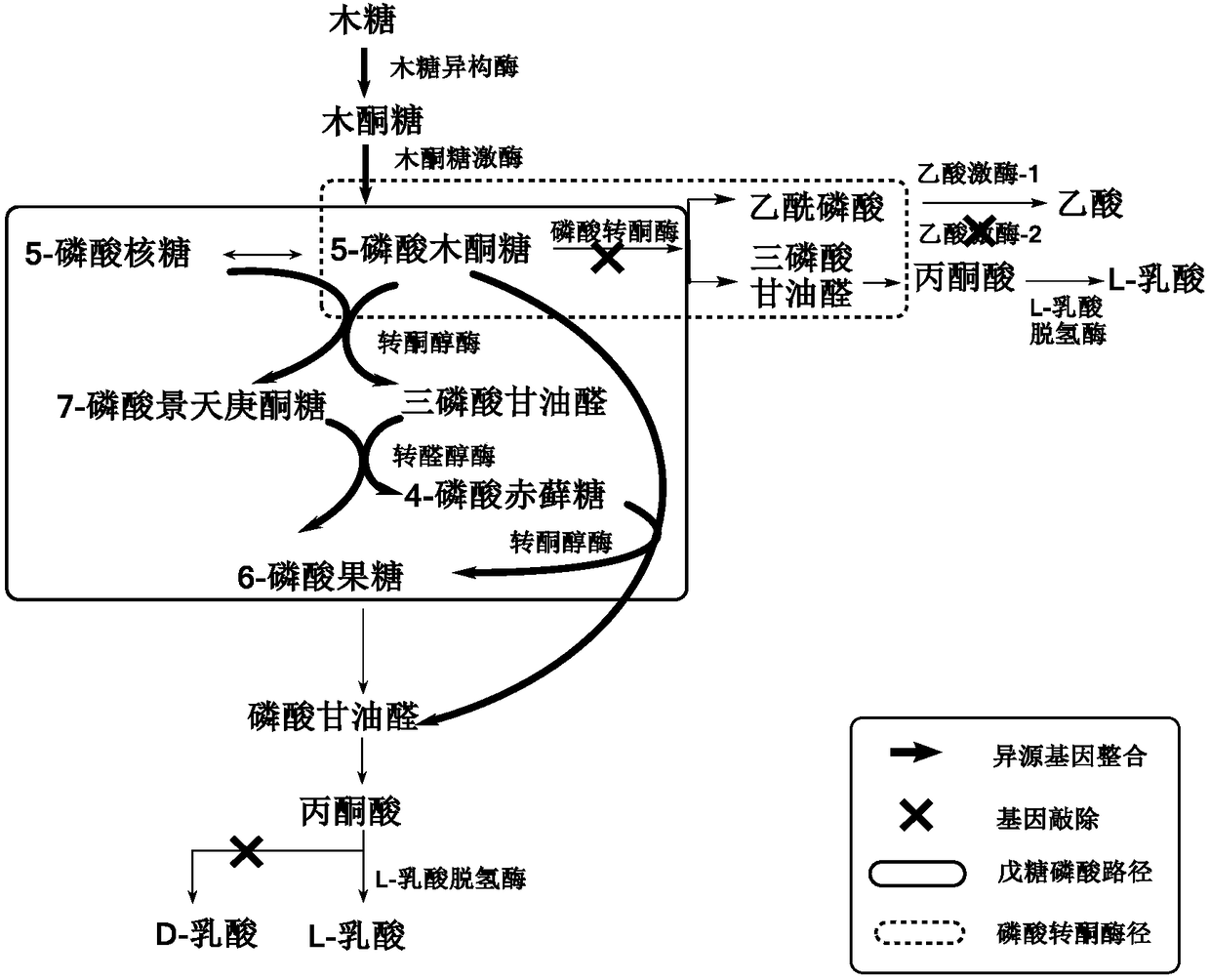
[0031] Implementation case 1: Implementation of xylose metabolism in P.acidilactici TY112.
[0032] (1) Construction of expression plasmid pMG36e-PldhD-xylAB_2911
[0033] First, using the P.acidilactici DSM20284 genome as a template, using xylAB_2911-F (SEQ ID NO:1) and xylAB_2911-R (SEQ ID NO:2) as primers to amplify to obtain xylAB_2911, using P.acidilactici TY112 as a template, and PldhD- F (SEQ ID NO: 3) and PldhD-R (SEQ ID NO: 4) were used as primers to amplify the promoter sequence PldhD about 300 bp upstream of the ldhD start codon, and then the expression box PldhD_xylAB_2911 was obtained by fusion PCR technology, and then passed EcoR I and Xba I double enzyme digestion, the P32 promoter of pMG36e itself was replaced with the expression cassette PldhD_xylAB_2911, and the expression plasmid pMG36e-PldhD_xylAB_2911 was obtained.
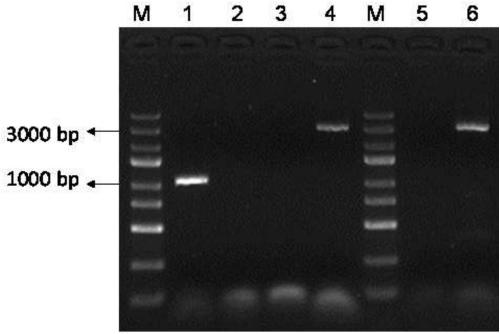
[0034] (2) Verification of xylose fermentation ability of recombinant strain P.acidilactici TY112 (pMG36e-PldhD_xylAB_2911)
[0035] The ex...
Embodiment example 2
[0036] Implementation Case 2: Blocking of the Phosphoketolase Pathway (PK Pathway)
[0037] (1) Construction of knockout plasmid pSET4E-Δpkt
[0038] Using the P. acidilactici TY112 genome as a template, using up-pkt-F (SEQ ID NO:5) and up-pkt-R (SEQ ID NO:6) as primers to amplify the upstream homology arm of about 1,000 bp of the pkt gene Fragment (up-pkt), using down-pkt-F (SEQ ID NO: 7) and down-pkt-R (SEQ ID NO: 8) as primers to amplify the downstream about 1,000bp homology arm fragment sequence of the pkt gene (down-pkt). The down-pkt fragment was inserted between the BamH I and Sac I sites of pSET4E, and then the up-pkt fragment was inserted between the Pst I and Sal I sites to obtain the plasmid pSET4E-Δpkt for pkt gene knockout.
[0039] (2) Knockout of phosphoketolase gene pkt
[0040] The knockout plasmid pSET4E-Δpkt was electrotransformed into P. acidilactici TY112 to obtain the recombinant strain P. acidilactici TY112 (pSET4E-Δpkt), and the recombinant strain wa...
Embodiment example 3
[0043] Implementation Case 3: Construction of the Pentose Phosphate Pathway (PP Pathway)
[0044] (1) Construction of integrated plasmid pSET4E-Δpkt::(tkt_tal)
[0045] Using the P.acidillactici TY112 genome as a template, using PldhD-F (SEQ ID NO:3) and PldhD-R (SEQ ID NO:4) as primers to amplify the promoter PldhD; using P.acidillactici DSM20284 as a template, design primers tkt_tal-F (SEQ ID NO: 11) and tkt_tal-R (SEQ ID NO: 12) were amplified to obtain tkt_tal. Then the cloned PldhD and tkt_tal were obtained by fusion PCR to obtain the expression cassette PldhD_tkt_tal, and inserted between Sal I and BamH I of the knockout plasmid pSET4E-Δpkt obtained in Case 2 to obtain the integrated plasmid pSET4E-Δpkt::(tkt_tal).
[0046] (2) Genomic integration of tkt_tal
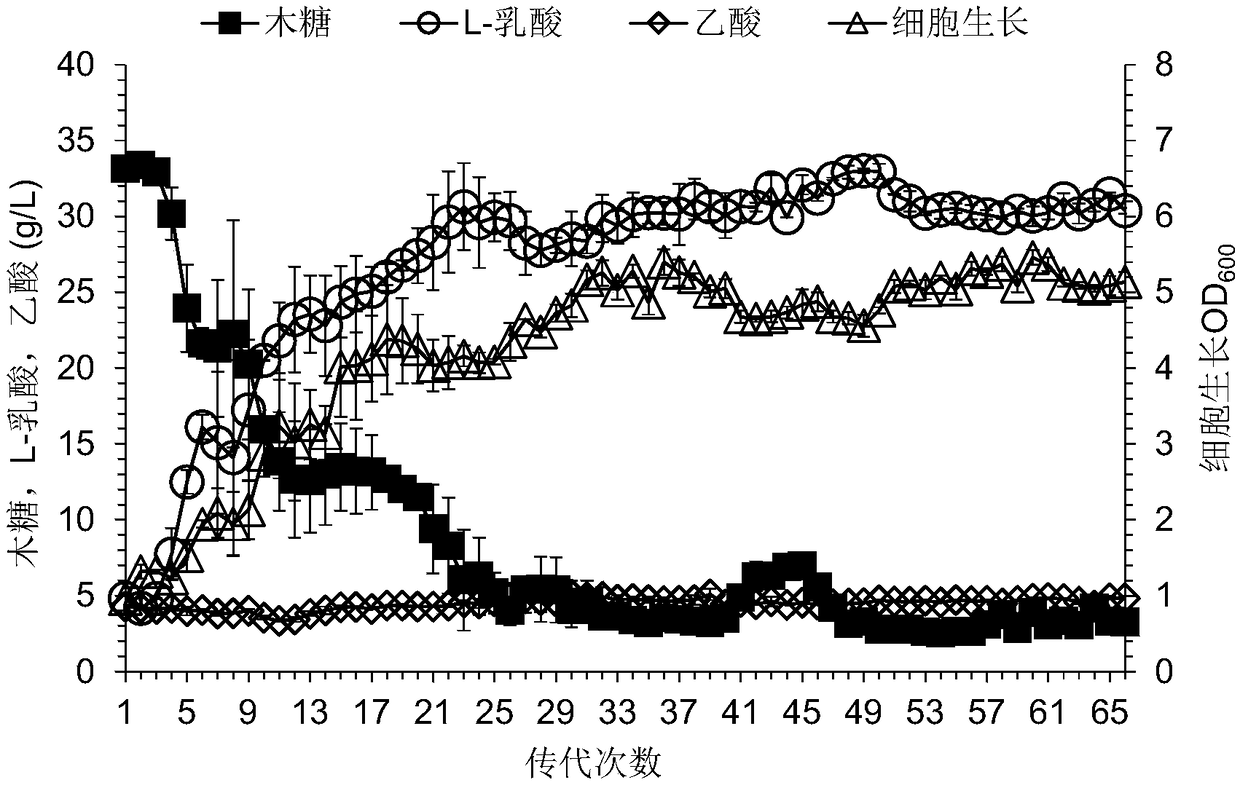
[0047] The integrated plasmid pSET4E-Δpkt::(tkt_tal) was electrotransformed into the engineering strain P. acidilactici TY112-Δpkt obtained in Case 2 to obtain the recombinant strain P. acidilactici TY112-Δpkt (pSE...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com