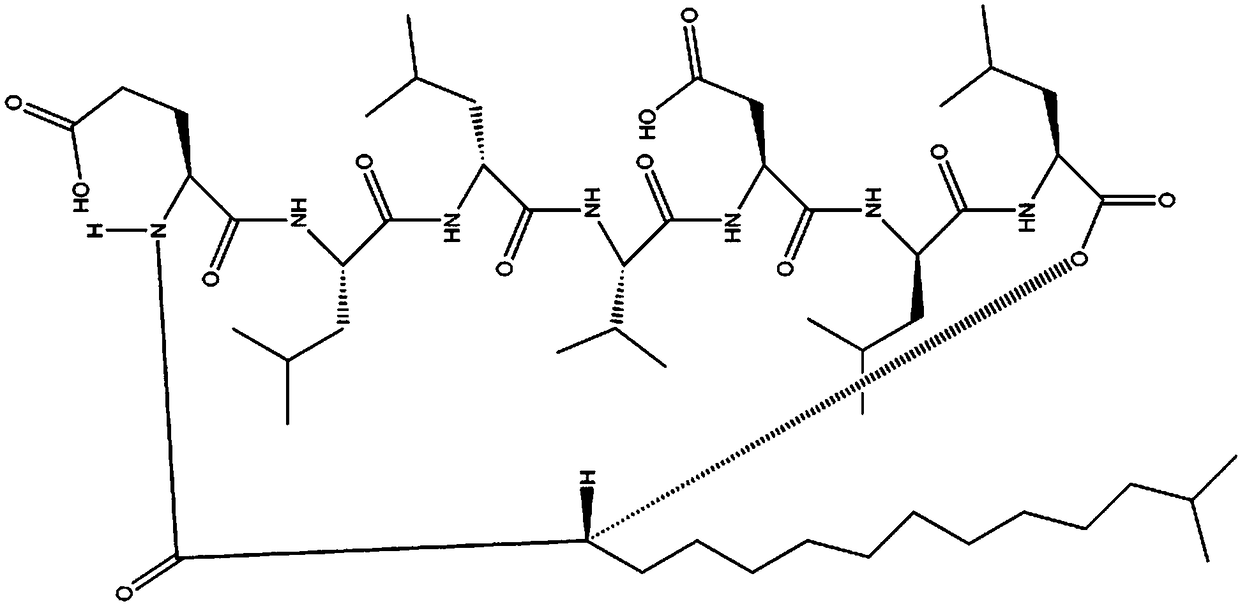
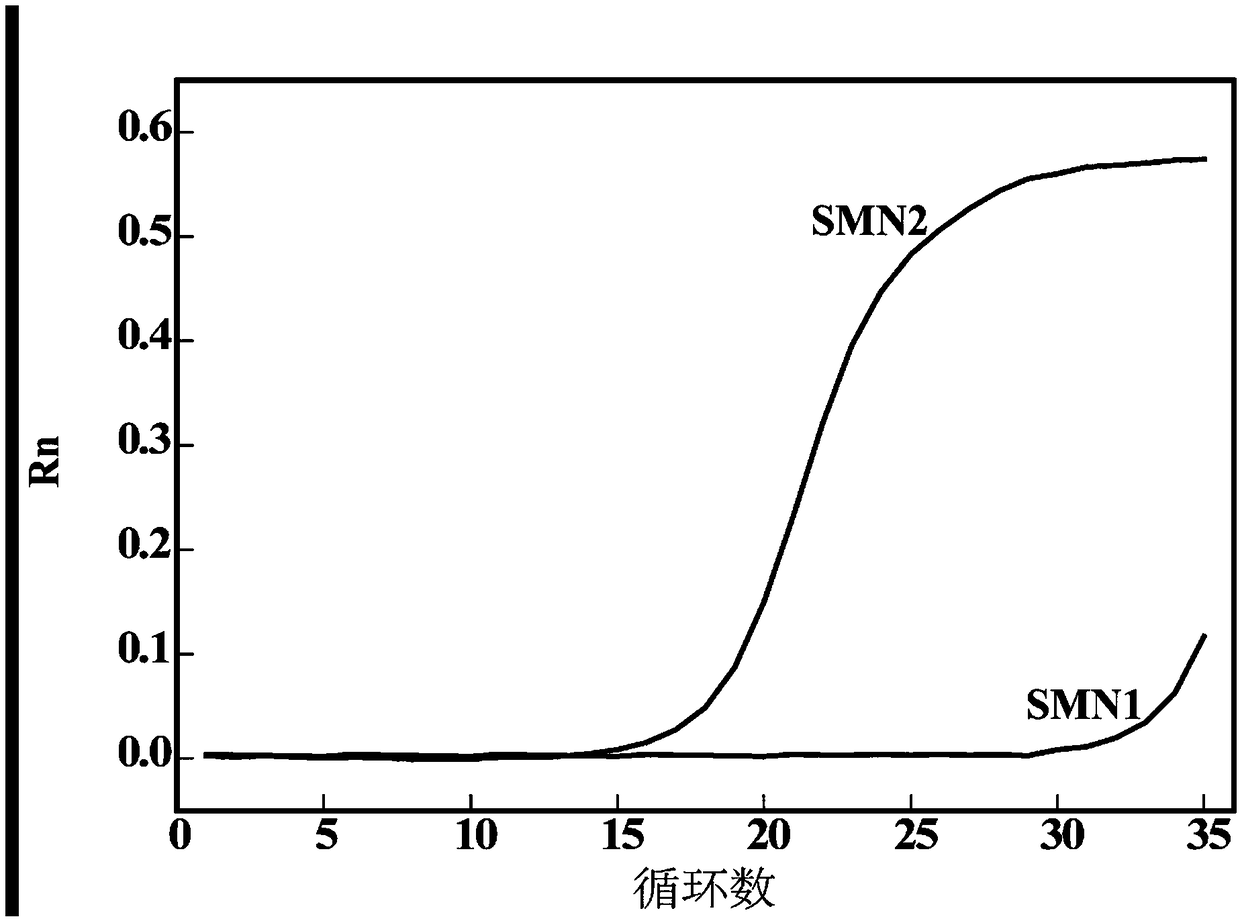
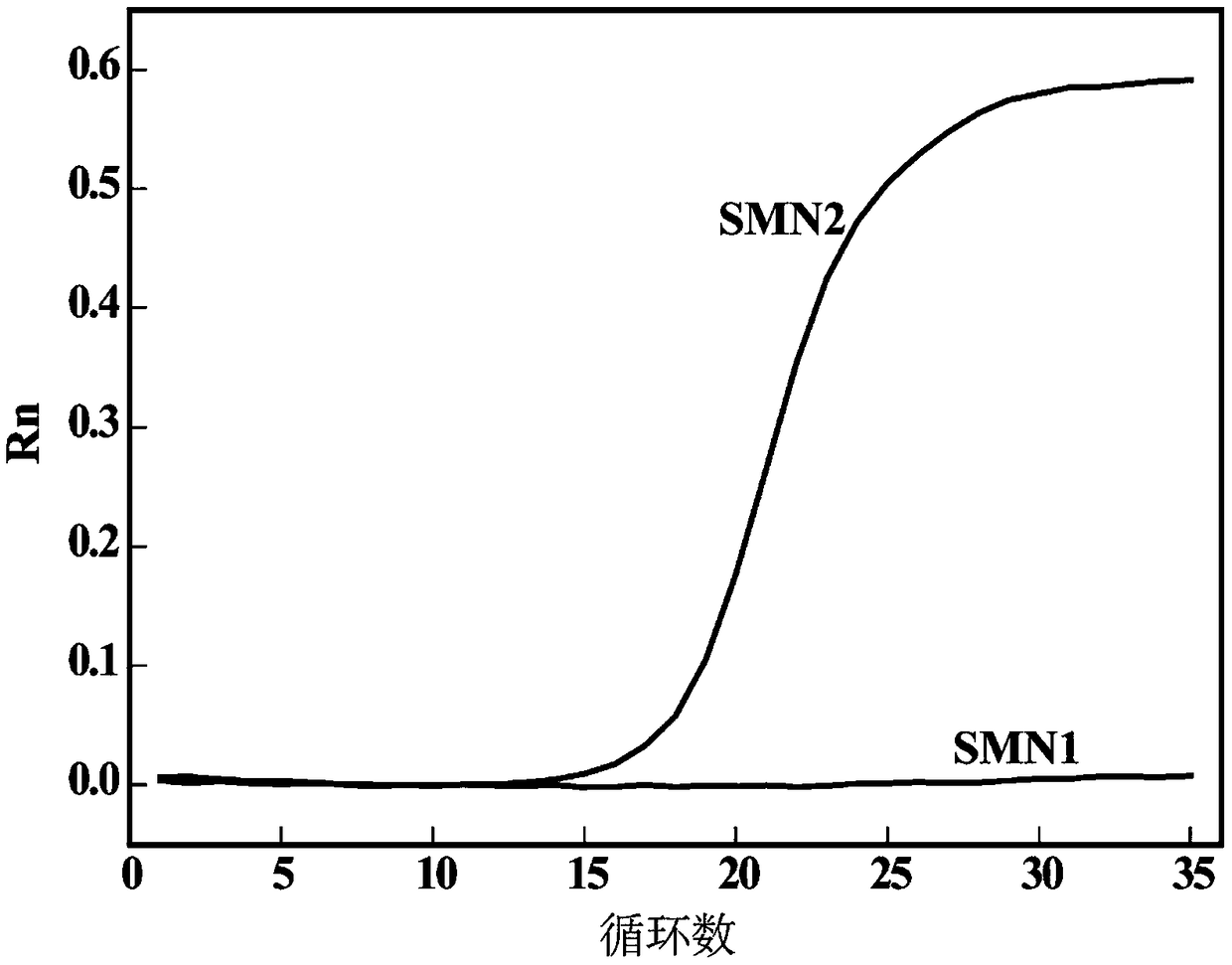
Detection method for number of multiple copies of SMN2 gene
A detection method and multi-copy technology, which are applied in the determination/inspection of microorganisms, biochemical equipment and methods, etc., can solve the problems of high cost, poor accuracy, and high requirements for sample purity, and achieve the effect of enhancing stability and improving accuracy.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0054] The present invention will be specifically introduced below in conjunction with the accompanying drawings and specific embodiments.
[0055] 1. To verify the influence of the amount of dNTPs and Surfactin on the detection effect of plasmid standards:
[0056] The method for detecting multiple copies of the SMN2 gene comprises the following steps:
[0057] step one,
[0058] Download the SMN1, SMN2 and reference gene RPP30 sequences from the UCSC website, compare the SMN1 and SMN2 gene sequences on the Clustal-X 1.8 software, and determine the difference between the two genes. The comparison results show that there are five The base difference is located in the No. 6 intron, No. 7 exon, No. 7 intron and No. 8 exon.
[0059] Design two specific upstream primers SMN2-F according to the 6th intron differential site of SMN2 0 and SMN2-F, the SMN2-F 0 Put the difference base at the last position of the 3' end of the primer; the SMN2-F covers the difference base at the 3' ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com