Construction method and application of annular RNA overexpression system of protoplast of moso bamboo
A protoplast and construction method technology, applied in the field of genetic engineering, can solve the problems of difficult to see the influence of circular RNA, great difficulty, and long time-consuming, and achieve the effect of saving economic costs, saving time, and stabilizing instantaneous transformation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Example 1 Extraction of Moso bamboo nucleic acid
[0040] (1) Material handling
[0041] Select bamboo shoots of 0.2 cm in the aboveground part of moso bamboo, cut the middle part with a scalpel, wrap it in tin foil, and put it in liquid nitrogen quickly. The material was ground using a high-throughput tissue grinder (QIAGEN TissueLyser II) and stored at -80°C.
[0042] (2) Extraction of Genomic DNA and Total RNA of Phyllostachys pubescens
[0043] Take 150 mg of the above ground sample, use Plant Genomic DNA Kit (TIANGEN, no. DP305, China) to extract the Genomic DNA of bamboo shoots; use RNAprep Pure Plant Kit (Polysaccharides&Polyphenolics-rich) (TIANGEN, no. DP441, China) to extract Total RNA. Total RNA is used for circular RNA verification, and DNA is used for vector construction and host gene amplification.
[0044] (3) RNA reverse transcription
[0045] Using PrimeScript ™ II 1st Strand cDNA Synthesis Kit (TaKaKa, no.6210A), 1 ug of Total RNA was reverse-tran...
Embodiment 2
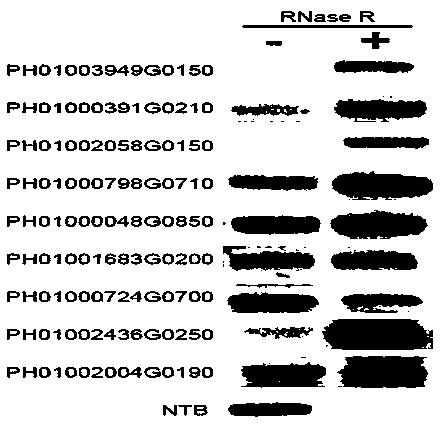
[0046] Example 2 Verification of circular RNA
[0047] According to the transcriptome data, nine circular RNA sequences were obtained, the sequences of which are shown in SEQ ID NO: 1 to SEQ ID NO: 9, and the "back-to-back" primers designed by the PRAPI software were used to verify the circular RNAs. The primer sequences are shown in Table 1.
[0048] 20 μg Total RNA (adjusted to 51 μL volume) was evenly divided into two tubes, one tube was treated with RNase R digestion, and the other tube was used as a control. Add 3 μL 10×RNase R Reaction Buffer, 1.5 μL RNase R (20U / uL) to the treatment tube; add 3 μL 10×RNase R Reaction Buffer, 1.5 μL RNase-freewater to the control tube. Incubate the treatment tube and the control tube in a water bath at 37 °C for 15 min, then add 30 μL of phenol-chloroform-isoamyl alcohol (25:24:1) to the two tubes to terminate the reaction, mix well, 4 °C, 13,000 g Centrifuge for 5 min. Transfer the supernatant to a new 1.5 mL RNase-free centrifuge tub...
Embodiment 3
[0050] Example 3 Construction of circular RNA overexpression recombinant plasmid
[0051] (1) Amplification of the target gene
[0052] The PH01000724G0700 (PH01000724:425581-426013) host gene was selected as the target gene. Design the 5' end of the target gene with attB- Specific primers for site adapters. The specific primer sequences are as follows:
[0053] The forward primer is: SEQ ID NO:28:
[0054] 5' GGGGACAAGTTTGTACAAAAAAGCAGGCTT CGTAAGTGGACGAAACACAAGAAG
[0055] AA-3' reverse primer is: SEQ ID NO:29:
[0056] 5' GGGGACCACTTTGTACAAGAAAGCTGGGTC CTTTTACCCCGTCAATCAAACCTTA-3'
[0057] The bold underlined part is attB recognition site.
[0058] Taking the Genomic DNA of Moso bamboo as a template, the above bands attB- 2.5 μL each for the forward and reverse primers of the adapter, 2×Unique HiQ TM Pfu Master Mix (No Dye) (Novogene, no. NHP007L, China) 25 μL, add ddH 2 0 to 50 μL system, 34 PCR cycles were performed at 98°C for 10 s, 58°C for 20 s, and 72°...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com