DNA molecular tile or nucleic acid nano-structure thereof, and applications thereof
A technology of DNA molecules and nanostructures, applied in recombinant DNA technology, DNA/RNA fragments, biochemical equipment and methods, etc., can solve problems such as limited Z-DNA
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1c64
[0056] The synthesis of embodiment 1c64nt, c84nt, c128nt
[0057] The 64nt, 84nt, and 128nt small circular DNAs used in the experiment were obtained by intramolecular circularization with T4 ligase. A total volume of 80 μL liquid containing 20nt splint DNA (4.5 μM) and 5’ phosphorylated DNA (3.5 μM), annealed from 95°C to room temperature within 2 hours, the splint strands will connect the 5’ phosphorylated DNA end to end, after annealing Add 10 μL of 10×T4 ligase buffer (660 mM Tris-HCl, 66 mM MgCl 2 , 100mM DTT, 1mM ATP), 10μL T4 ligase (300U / μL), the total volume of 100μL was reacted in a 16℃ water bath for 16h. Heat at 95°C for 5min, extract and cool in an ice-water bath, add 10μL of 10× exonuclease I buffer, 10μL of exonuclease I (5U / μL), incubate in a 37°C water bath for 30min, and excise the remaining linear DNA. Then use 8-12% denaturing polyacrylamide gel electrophoresis (PAGE) to separate. Cut the target band, crush it, add twice the mass of deionized water, mix a...
Embodiment 2
[0058] Example 2 HJ@c64nt, cDAO@c64nt molecular tiles and their application
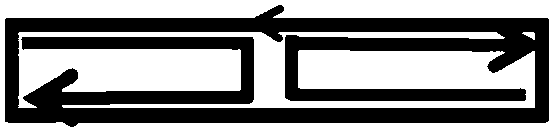
[0059] The molecular tile structure of HJ@c64nt is as follows figure 1 As shown, the black rectangular line seamlessly connected in the middle is a covalently linked 64nt DNA small circle (c64nt), and the direction pointed by the arrow represents the direction of the DNA from 5' to 3' (except for the description of the arrow below point out, representing the direction of DNA); the two dark gray linear DNA strands form a HJ cross junction in the middle of the c64nt small circle to divide c64nt into two equal parts, and the two single strands are balanced and complementary to the sequences on both sides of the HJ respectively, which is used here Line segments and arrows are shown to represent DNA strands and their orientation.
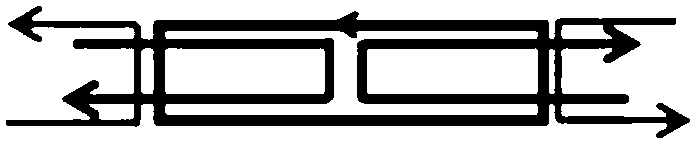
[0060] The molecular tile structure of cDAO@c64nt is as follows figure 2 As shown, in HJ@c64nt, the four heads of the two back-turned dark gray linear single strands protrud...
Embodiment 3
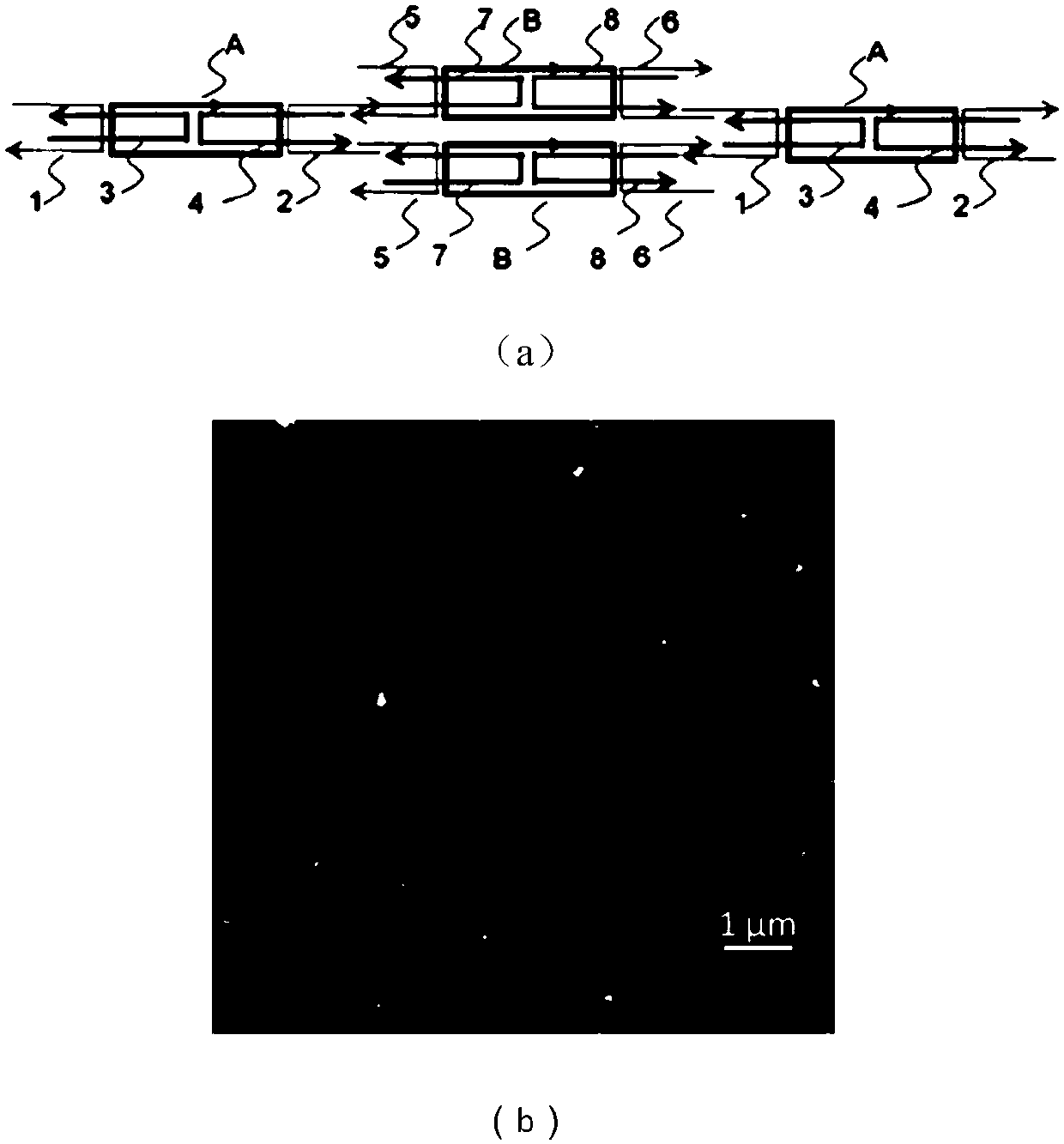
[0063] Example 3 cDAO@c64nt-E (-E=4 semi-helical coils) 2D array connected by 21bp and its AFM image
[0064] Such as image 3 As shown in (a), the cDAO@c64nt composed of two loops of different sequences A and B are connected. The helical directions of the two loops of A and B are the same, and a connection of 21bp (-E=4 half-helical turns) is used. That is, four overhanging 8bp cDAO@c64nt molecular tiles with 5nt cohesive ends are used for assembly to form a two-dimensional array structure.
[0065] The experimental steps are as follows: the A ring, the B ring are mixed with the four straight chains constituting the stable molecular tiles in a mixture containing 12.5mM Mg(Ac) 2 In 1×TAE (40 mM Tris, 40 mM HAc, 1 mM EDTA, pH=8.0) solution, the final concentration of each DNA strand was 0.5 μM, and the total volume was 20 μL. Then, a 2D array of cDAO@c64nt-E was prepared by using the step-by-step method or the one-pot method in the specific embodiment.
[0066] image 3 The...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com