Male specific DNA marker of oplegnathus punctatus and method for identifying hereditary sex
A technology of zebra sea bream and gender, which is applied in the field of male-specific DNA marker and genetic sex identification of zebra sea bream, can solve problems such as unreported, and achieve the effects of shortening time, saving detection time and cost.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Example 1 Screening and verification of male-specific DNA fragments of porpoise
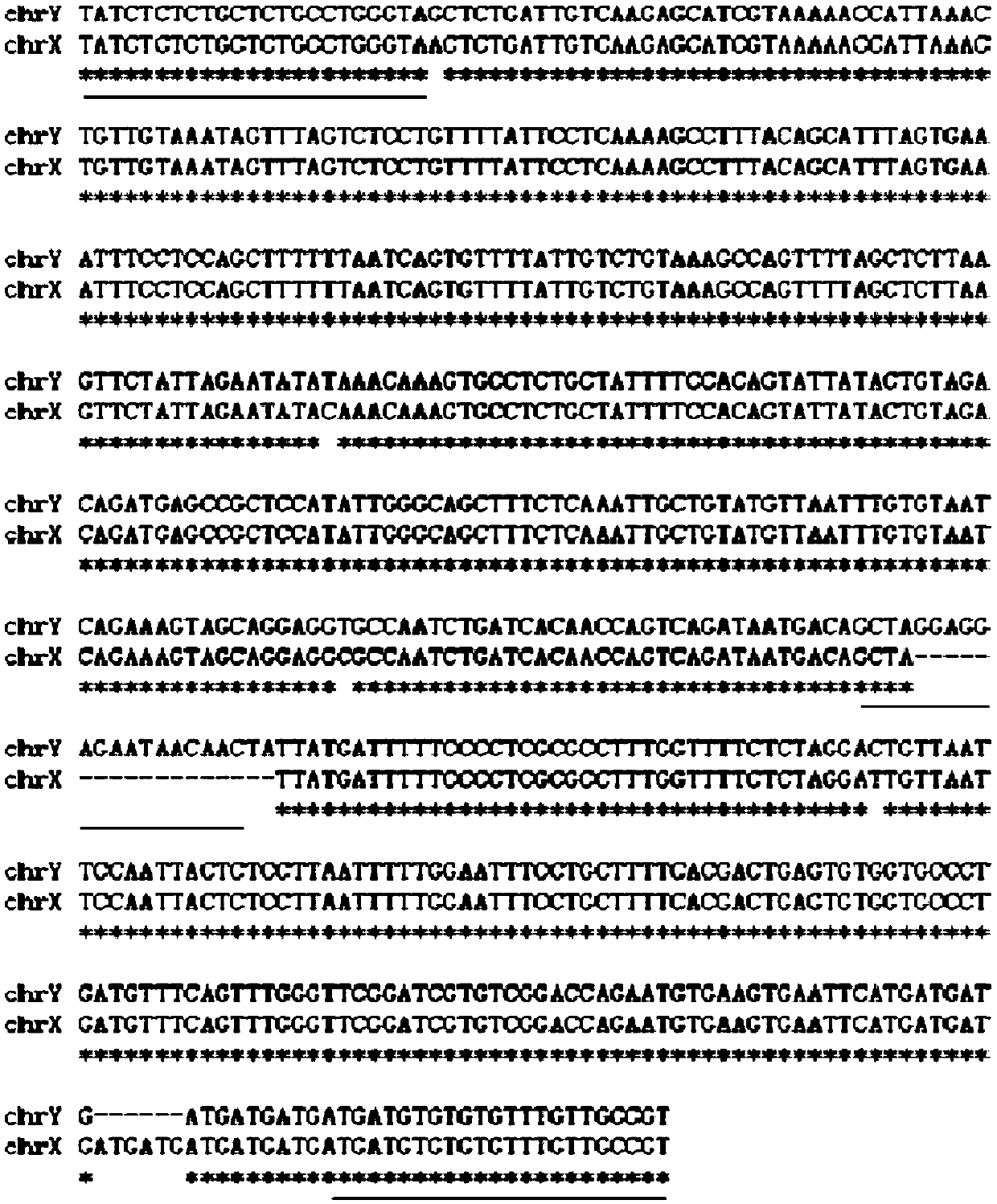
[0022] The discovery of male-specific DNA fragments: the sex chromosome-linked DNA fragment sequences used in the present invention are derived from the whole genome sequencing results of the grouper sea bream (unpublished Published), bioinformatics analysis of the genome sequence of P. chinensis screened out two homologous and different DNA fragments of X and Y chromosomes. Two homologous differential DNA fragments of X (X1 or X2) and Y chromosomes were found. The DNA fragment on the Y chromosome was 573bp long, named SEQchrY, and its sequence was SEQ ID NO: 1:
[0023] TATCTCTCTGCTCTGCCTGGGTAGCTCTGATTGTCAAGAGCATCGTAAA AACCATTAAACTGTTGTAAATAGTTTAGTCTCCTGTTTTATTCCTCAAAAGC CTTTACAGCATTTAGTGAAATTTCCTCCAGCTTTTTTAATCAGTGTTTTATTG TCTGTAAAGCCAGTTTTAGCTCTTAAGTTCTATTAGAATATATAAACAAAGT GCCTCTGCTATTTTCCACAGTATTATACTGTAGACAGATGAGCCGCTCCATA TTGGGCAGCTTTCTCAAATTGCTGTATGTTAATTTGTGTAATCAGAAAGTAG CAGGAGGT...
Embodiment 2
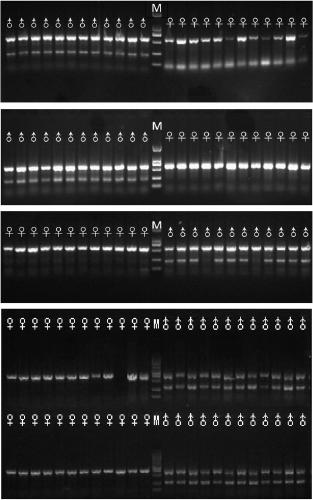
[0028] Example 2: Establishment and application of genetic sex identification technology for porpoise
[0029] 1. Genetic sex identification of porpoise under laboratory conditions
[0030] DNA extraction: Use the marine organisms DNA extraction kit (Tiangen) to extract the DNA of the fin ray of the grouper, use 1% agarose gel electrophoresis to identify its integrity, measure its OD value with a UV spectrophotometer, and adjust the DNA concentration to 50ng / μL, and stored at -20°C until use.
[0031] PCR identification: using the sex-specific DNA fragment primers SEQ XY: 1, SEQ XY: 2, and SEQ Y: 3 of the grouper to detect the genetic sex of the grouper by PCR. The PCR reaction system is 15 μL, including 1.5 μL of 10× buffer, 0.8 μL of dNTP, 0.3 μL of each upstream primer, 0.6 μL of downstream primer, 1 μL of template DNA (50ng / μL), 11.0 μL of ddH2O, and 0.1 μL of rTaq enzyme. PCR amplification program: 95°C for 5min; 30 cycles of 95°C for 30s, 60°C for 30s, and 72°C for 30...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com