Traceless double-target genome editing system
A genome editing, dual-target technology, applied in the field of genome editing, can solve the problems of resistance gene removal, difficulty, and insufficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment example 1
[0044] Implementation case 1. Simultaneously gal K (PAM dependent) and lac Z gene (PAM-independent) point mutation
[0045] Escherichia coli E. coli W3110
[0046] Table 1 Primers and related sequences required for double point mutation
[0047]
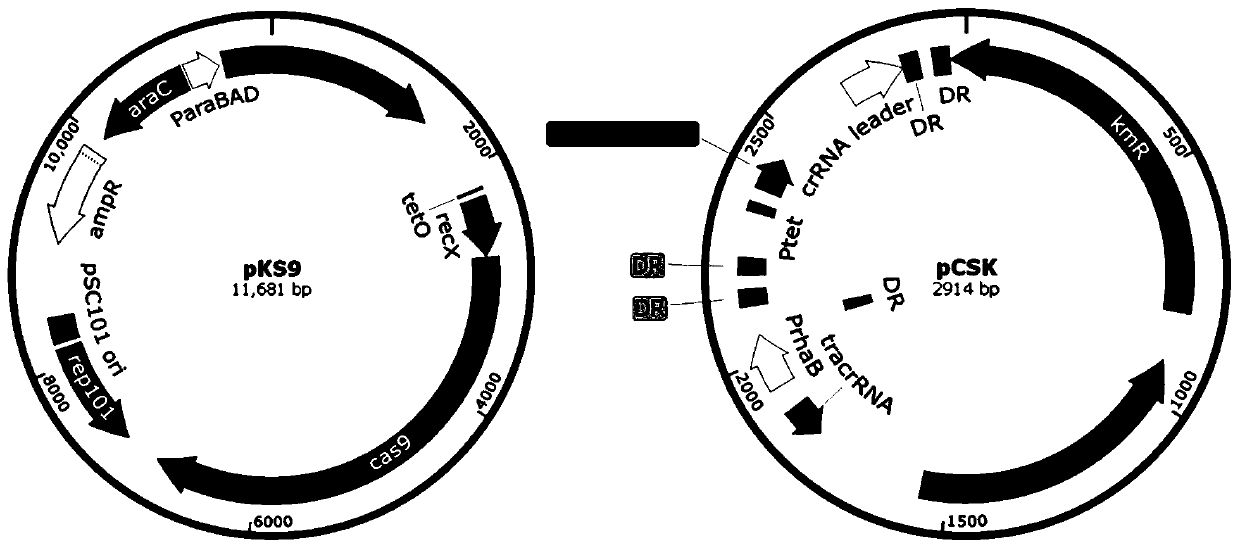
[0048] (1) Transformation of pCSK plasmid: Use the primer sequences in Table 1 to carry out pCSK plasmid gal K and lac Z Addition of target sequence gRNA: the specific method is as follows:
[0049] gal The K sgRNA was added behind the tetracycline promoter using the USER-LIC cloning method. The specific step is to combine two new target gal The K sgRNA complementary single strand was treated in a water bath at 95°C for 5 min and then annealed overnight, then mixed with the pCSK product amplified by the pCSK primers, treated with USER enzyme and then transferred to DH5a for identification to complete the pCSK plasmid backbone and gal K sgRNA binding.
[0050] lac The position of the Z crRNA target is behind the...
Embodiment example 2
[0055] Implementation case two, at the same time gal K gene and ATP-dependent RNA helicase dbp A gene replacement
[0056] Adopt bacterial strain DH5a, adopt table 2 primer and sequence:
[0057] Table 2 Simultaneous gene replacement and gene insertion primers and sequences
[0058]
[0059] The research method is the same as the implementation case 1, of which about 700bp gal K gene was replaced by 1247bp fragment, 1000bp dbpA gene is 560bp lacZ ' DNA fragment substitution. because lac The insertion of Z' can realize α-complementation in the DH5a genome, so that the mutants can form blue colonies on the LB plate containing IPTG and XGal. for gal The identification after the K gene replacement still uses the MacConkey red and white spot method for preliminary screening. In addition, the homologous recombination sequence used this time is a double-stranded DNA PCR product with 40bp homology arms on both sides. gal The K. del700bp.HP primer produces a fragme...
Embodiment example 3
[0062] Implementation case 3. Simultaneous lactosinase gal K gene replacement and β-galactosidase lac Z gene point mutation
[0063] Strains used: E. coli W3110, using primers and DNA sequences from Table 1 lac Part Z and Table 2 of gal K part composition.
[0064] The method adopted is the same as that of the implementation cases 1 and 2.
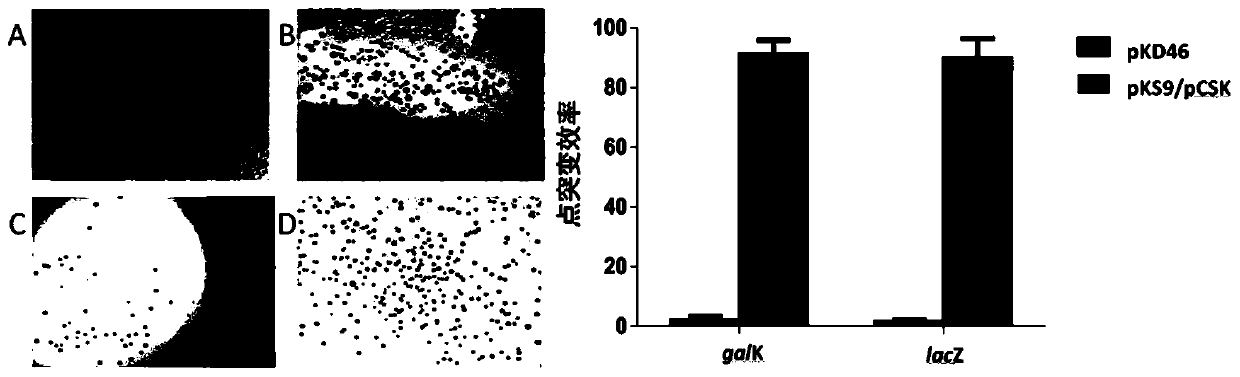
[0065] experiment result shows( Figure 8 ): gal The mutation efficiency of K is 61.4%, lac The mutation efficiency of Z was 95.1%.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com