Method for enriching and screening a target substance, such as cells, from sample
A technology in a target or sample, used in a device for bacteria or biomolecules to enrich and screen targets such as cells, which can solve the problems of small contact area and low capture rate of the device, and achieve improved capture rate, increased probability, and improved The effect of work efficiency and application range
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
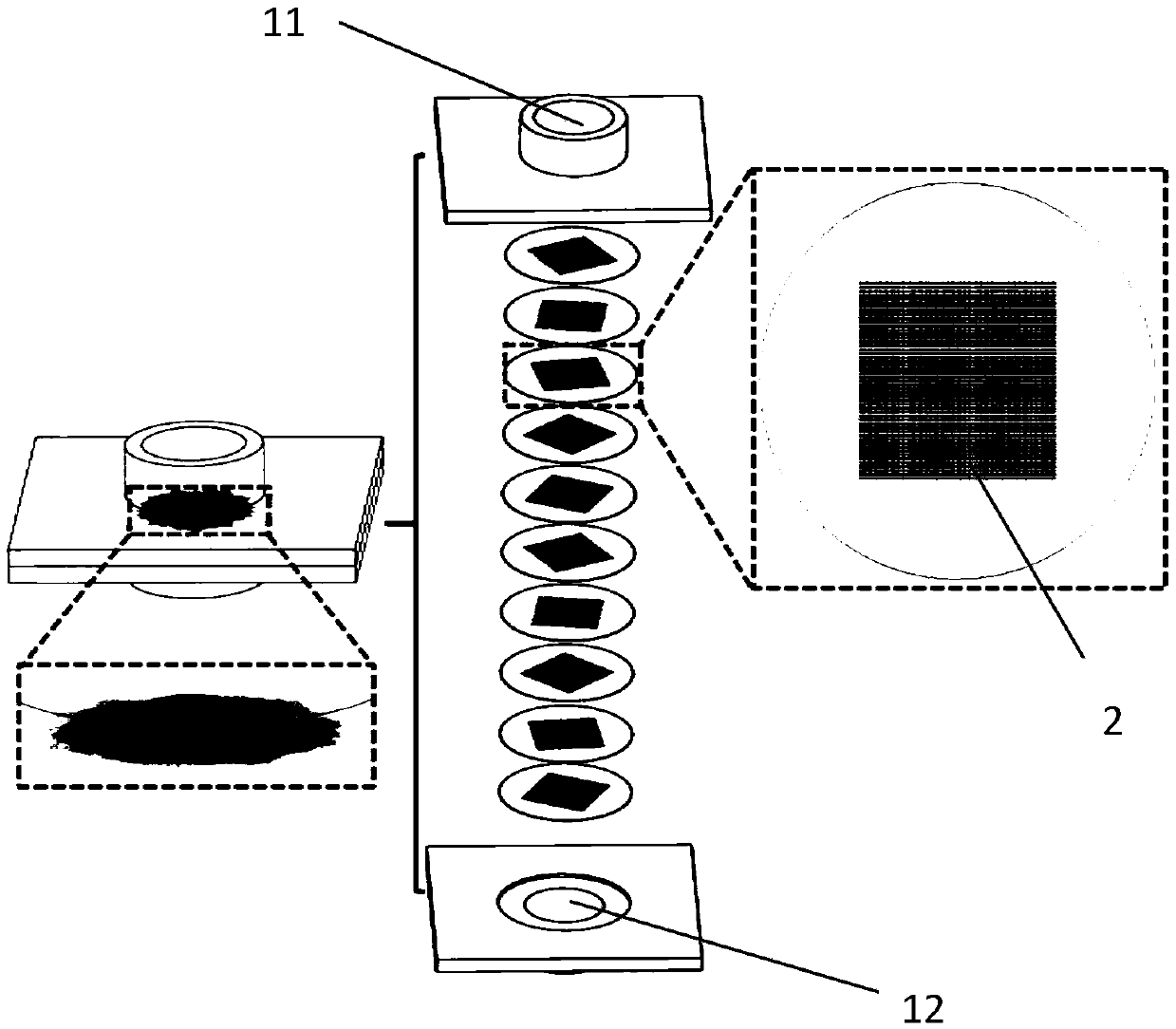
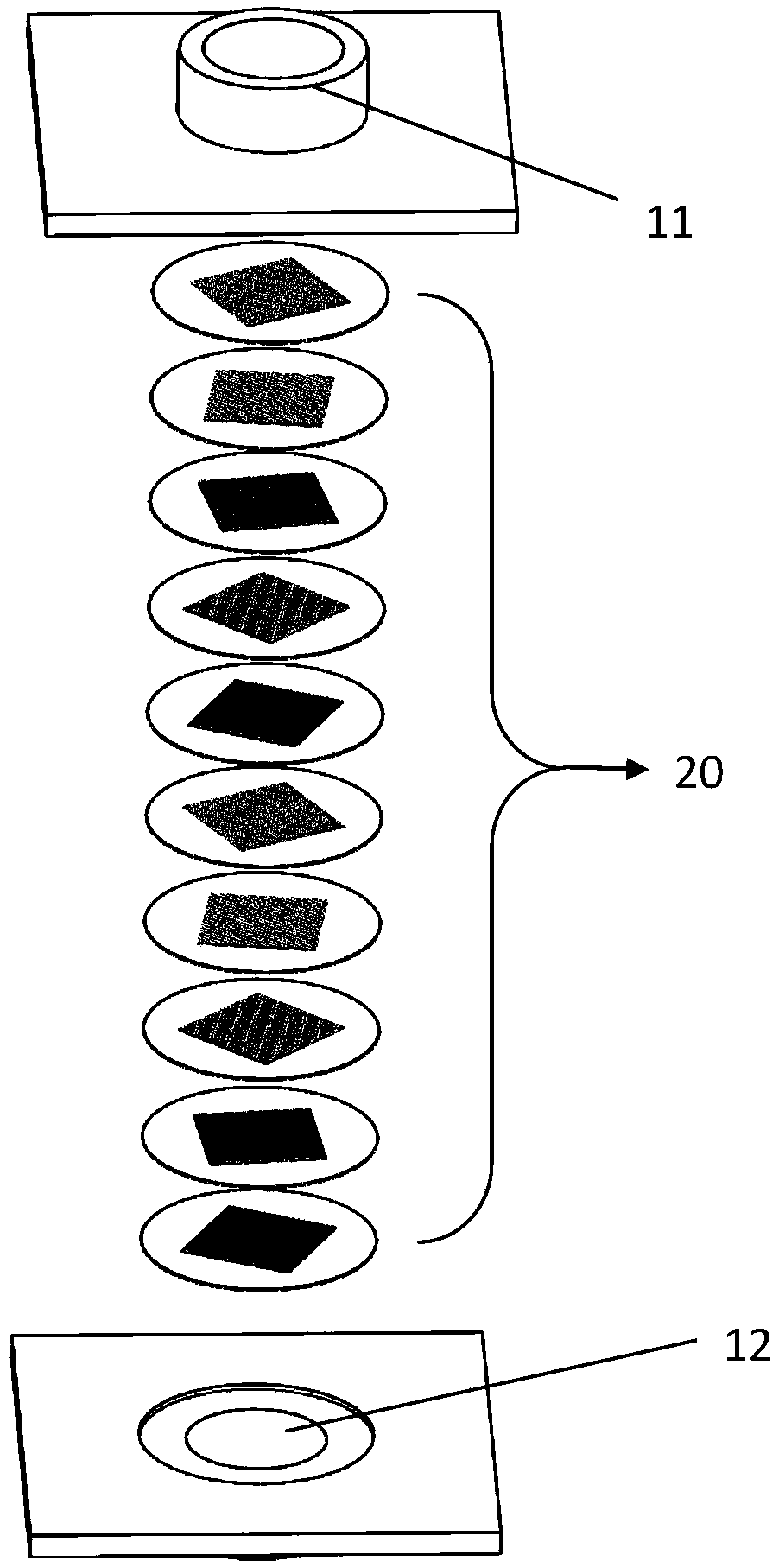
[0059] In one embodiment, the rotation angle parameter of the multi-channel network matrix 20 is expressed as [0:9:81], which means that the rotation angle of the multi-channel network matrix 20 relative to the upper multi-channel network matrix 20 is 9°. The number of the network-like bases 20 is ten. According to another embodiment, the rotation angle parameter of the multi-channel network matrix 20 is expressed as [0:180], which means that the multi-channel network matrix 20 is rotated 180° relative to the upper multi-channel network matrix 20, and the multi-channel network matrix 20 is rotated by 180°. The number of the shaped bases 20 is two.
[0060] The specific specifications of the mesh cable 23 and the screen hole 22 are mainly described below.
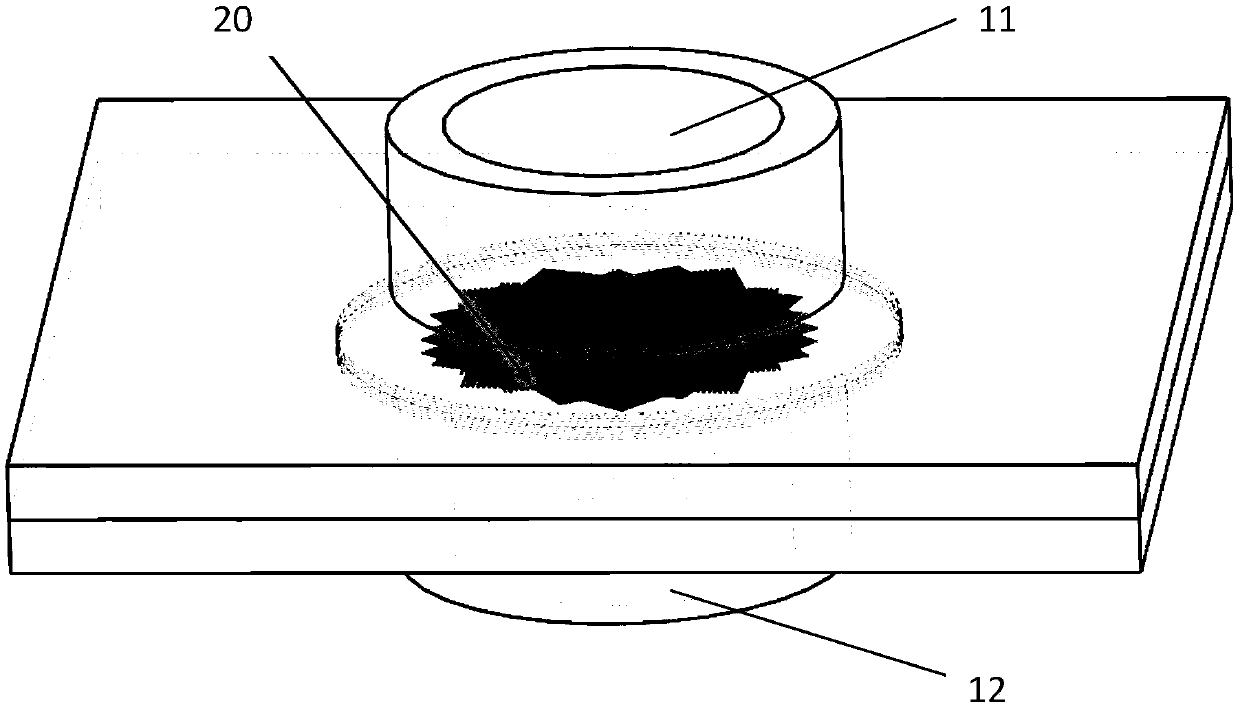
[0061] The shape of the cross section of the screen hole 22 is one or more of square, rectangle, triangle, polygon, circle, parallelogram or trapezoid. In this article, as attached Image 6 As shown, the thickness D of th...
Embodiment 1
[0076] An enrichment screening mechanism, wherein the angle rotation parameter of the plurality of porous network substrates 20 is [0:36:324], that is, the rotation angle of the lower layer porous network substrate 20 relative to the upper layer porous network substrate 20 is 36°, and there are 10 porous mesh bases 20 in total.
[0077] The distance between adjacent multi-channel mesh substrates 20 is 1 mm, the thickness of the mesh wire 23 is 22 μm, the thickness is 22 μm, the cross section of the sieve hole 22 is square, and the cross-sectional area of the sieve hole 22 is 54×54 μm 2 .
[0078] The capture objects on each porous network substrate 20 are EpCAM antibodies, and the method for connecting the capture objects on the porous network substrate 20 includes the following steps:
[0079] (1) Take 4.6 microliters of Traut's reagent solution (0.2 mg / ml), 5 microliters of EPCAM antibody (Cat#ab32392, Abcam), 40.4 microliters of PBS-EDTA (2.5 mmol, pH8.0) and mix the thr...
Embodiment 2
[0092] It is basically the same as Example 1, except that the angle rotation parameter of the plurality of porous network substrates 20 is [0:9:81], that is, the lower porous network substrate 20 is relative to the upper porous network substrate 20. The angle of rotation is 9°.
[0093] The number of lung cancer cells retained in this example was 5.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Roughness | aaaaa | aaaaa |
| Thickness | aaaaa | aaaaa |
| Roughness | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com