One-step washing magnetic bead method blood dna extraction kit
The technology of a kit and washing solution is applied in the field of magnetic bead method blood DNA extraction kit, which can solve the problems such as easy pollution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Embodiment 1 Reagent preparation and basic extraction process
[0049] Prepare extraction reagent 1 according to the following recipe:
[0050] Lysis buffer: Tris 60mM, EDTA 15mM, NaCl 0.6M, SDS 0.3%, CHAPS 0.2%, NLS 7%, GuHCl72%, sodium laurate 3%, NH 4 Cl 0.2%, mercaptoethanol 1.0%, pH 7.1;
[0051] Binding solution: isopropanol 70%, PEG 8000 8%, Brij 58 3%, pH 7.0;
[0052] Proteinase K solution: proteinase K 20mg / ml;
[0053] Washing solution: GuHCl 7M, Tris 15mM, ethanol 50%, sodium bicarbonate 40mM, pH 9.0;
[0054] Elution buffer: Tris 20mM, EDTA 0.2mM, pH 8.5.
[0055] Control reagent 2 was prepared according to the formula of reagent 1 (replace sodium bicarbonate with sodium acetate 10mM)
[0056] Control reagent 3 was prepared according to the formula of reagent 1 (without CHAPS, with NaDC 0.6%)
[0057] Follow the steps below for DNA extraction:
[0058] (1) Take 200ul of fresh blood into a 1.5ml centrifuge tube, then add a total of 400ul of a 1:1 mixt...
Embodiment 2
[0063] Example 2 Validation of the effect of the kit formula of the present application
[0064] 1 The role of sodium bicarbonate and DPTA
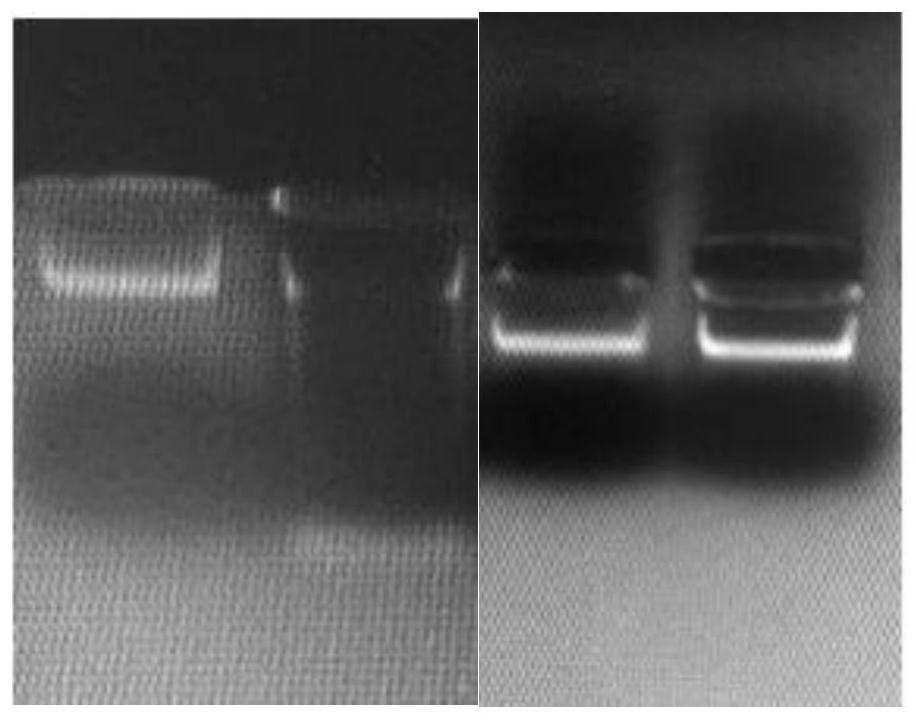
[0065] Using reagents 1-3 and DNA extraction method in Example 1 to extract the same blood sample, use Qubit® (based on fluorescent dye method) detector and matching kit to detect the concentration of extracted DNA and detect ODA260 / 280, the results as follows:
[0066]
[0067] Repeat results in multiple samples were similar. We speculate that the acid-base / charge conditions in the washing solution have a great influence on the washing effect of mucin hydrolyzate. Among the various formulas in the experiment, only slightly alkaline sodium bicarbonate can effectively remove protein without affecting DNA stability. The effect; the addition of zwitterionic surfactant is conducive to fully lysing the lysed protein and making it easier to remove the protein during washing.
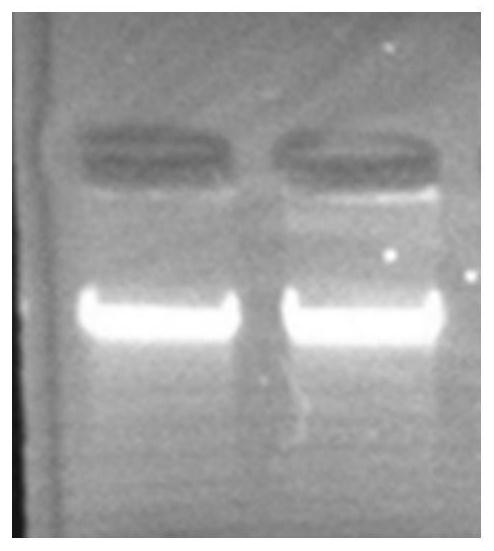
[0068] Comparison between the one-step washing kit of this app...
Embodiment 3
[0074] Example 3 Use the kit of this application for actual detection
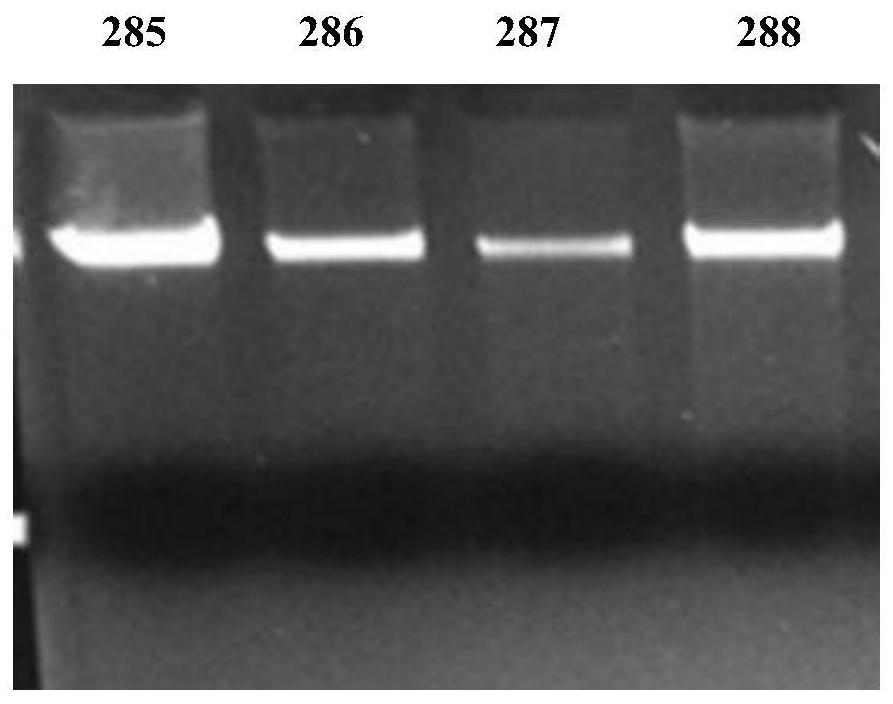
[0075] Use the kit of this application to extract 4 pairs of DNA from the blood of 8 subjects (collected synchronously with blood samples) for STR locus typing in paternity testing (Promega Power Plex16, 16 sites, ABI9700 PCR instrument), and Compared with the typing done by the DNA extracted from the blood, the typing situation of all 128 loci of the 8 test subjects is exactly the same. It is preliminarily proved that the sample extracted by the kit of the present application can be used for subsequent molecular biology detection.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com