Absolute quantification method for bacterial flora and application of absolute quantification method in Chinese liquor fermentation process
An absolute quantitative, liquor technology, applied in the fields of fermentation engineering and biology, it can solve the problems of interference, difficulty in finding, and inconvenient operation of bacterial flora structure sequencing results.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0068] Example 1: Design of Lactobacillus acetotolerans-specific primers
[0069] (1) Use the Blast function of the National Center for Biotechnology Information (NCBI) website (https: / / blast.ncbi.nlm.nih.gov / Blast.cgi) to manually screen from the whole genome of Lactobacillus acetotolerans (AP014808.1) The specific gene WP_082137158.1 (NCBI accession number) is functionally annotated as a transcriptional regulator. According to the comparison results, only the genome sequence of Lactobacillus acetotolerans (AP014808.1) has a 100% similarity, indicating that the nucleotide sequence of the transcriptional regulator is a highly specific sequence of Lactobacillus acetotolerans.
[0070] (2) Use Batchprimer online software to design specific primers. The parameters are set as follows: primer length 18-27bp, optimal 20bp; annealing temperature 57-62°C, optimal 60°C; GC content 40-60%, optimal 50% ; The temperature difference between the primer pair is set to be less than 5°C; the ...
Embodiment 2
[0074] Example 2: Design of Lactobacillus sp.-specific primers
[0075] (1) Since Lactobacillus sp. is an uncultivated microorganism, the only 16S rRNA differential region sequence (GenBank: KU674948.1) was selected for specific primer design. The differential sequence was obtained by comparing the following sequences: 1) According to the Blast comparison To the result (similarity is shown in parentheses after the bacterial species name), select the 16S rRNA sequence of the strain with high similarity: Lactobacillus caviae (90%), Lactobacillus fructivorans (90%), Lactobacillushomohiochii (90%), Lactobacillus ixorae (89%) ), Lactobacillus reuteri (89%), Lactobacillus vespulae (89%), Lactobacillus ozensis (88%); 2) Lactobacillus brevis (88%) and Lactobacillus acetotolerans (87%) that appear frequently in the liquor brewing system. The screening of the 16S rRNA nucleic acid sequence difference region was completed by using the DNAMAN bioinformatics software to compare with the 16...
Embodiment 3
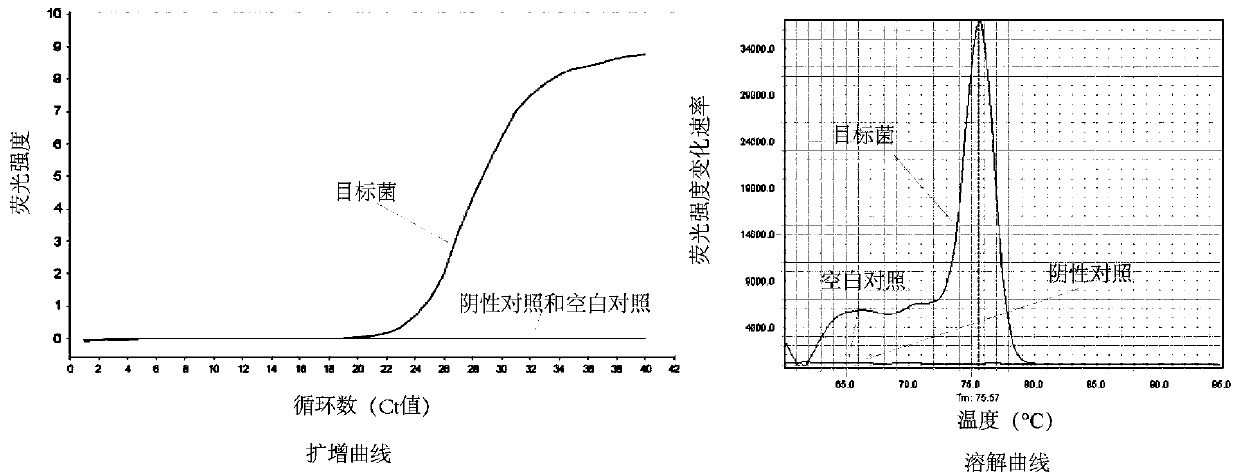
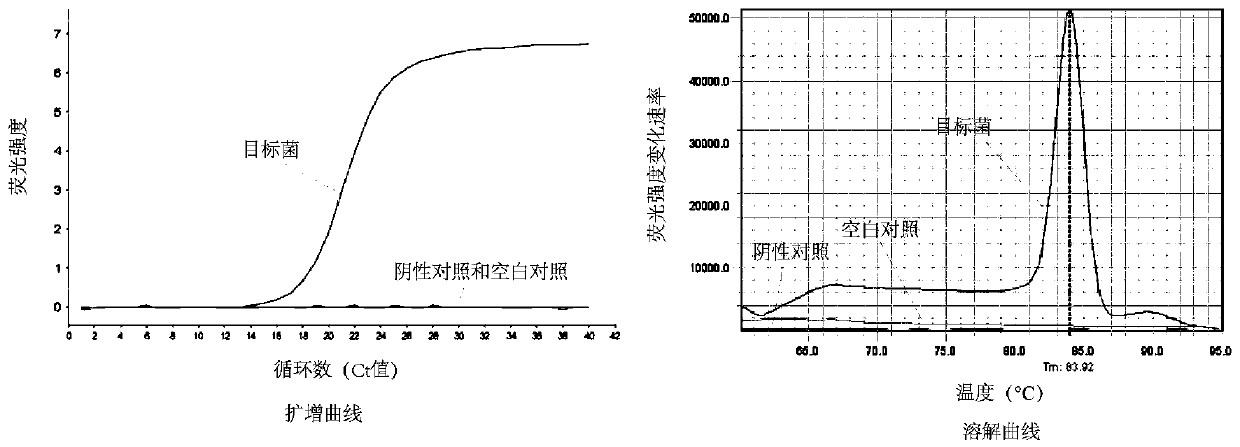
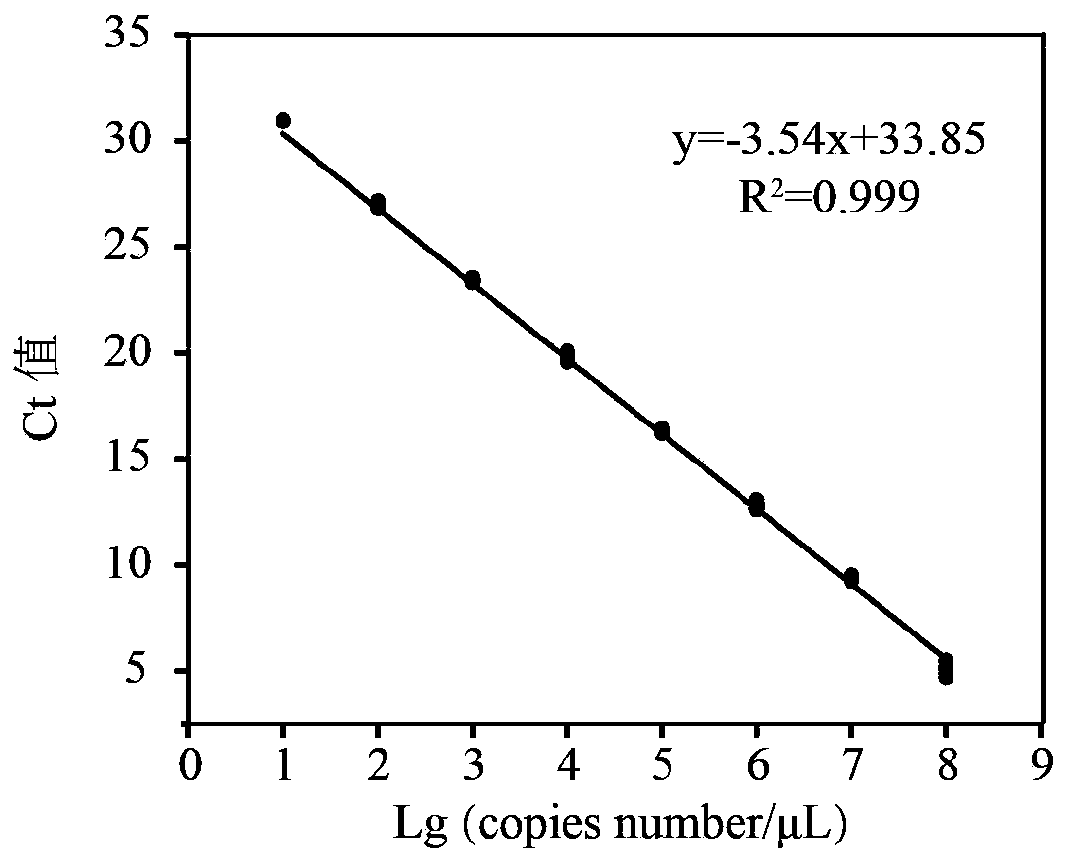
[0079] Example 3: Validation of Lactobacillus acetotolerans-specific primers
[0080](1) Select a target Lactobacillus acetotolerans for qPCR, and the negative control is 36 microorganisms from mouse intestine, liquor Xiaoqu, Daqu, commercial starter, yogurt, and silage: Lactobacillus buchneri, Lactobacillus dioilvorans, Lactobacillus brevis, Lactobacilluscrustorum, Lactobacillusplantarum ,Lactobacillus harbinensis,Lactobacillusacidiliscis,Pediococcus ethanolidurans,Pediococcus acidilactici,Pediococcuspentosaceus,Lactobacillus murinus,Lactobacillus curvatus,Lactobacillus casei,Lactobacillus reuteri,Lactobacilluspanis,Lactobacillusfermentum,Lactobacillusjohnsonii,Lactobacillus delbrueckii,Lactococcus lactis,Weissella confusa,Weissella paramesenteroides,Weissellaviridescens,Leuconostoc citreum,Leuconostoc lactis, Leuconostoc mesenteroides, Leuconostocpseudomesenteroides, Enterococcus italicus, Enterococcus lactis, Enterococcusfaecalis, Bacillus coagulans, Bacillus licheniformis, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com