Genetically engineered bacterium for expressing high-activity ice nucleation protein and construction method of genetically engineered bacterium
A technology of genetically engineered bacteria and ice nucleoprotein, applied in the field of genetically engineered bacteria expressing highly active ice nucleoprotein and its construction, can solve the problems of complex genetic background and limit the maximum utilization of ice nucleoprotein, and achieve clear genetic background, Improve the activity and efficiency of ice nucleoprotein
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] The selection and optimization of the starting bacterial strain of embodiment 1
[0026] Although Escherichia coli is an conditional pathogen, its inactivated strain is not pathogenic, and the inactivated strain can also ensure the activity of ice nucleoprotein, and the genetic background of Escherichia coli is clear, and the genetic modification is highly safe and operable. And efficient, so Escherichia coli was selected as the starting strain of the ice nucleoprotein engineering bacteria.
[0027] Wild-type Escherichia coli MG1655, DH5α and BW25113 can all be used as starting strains.
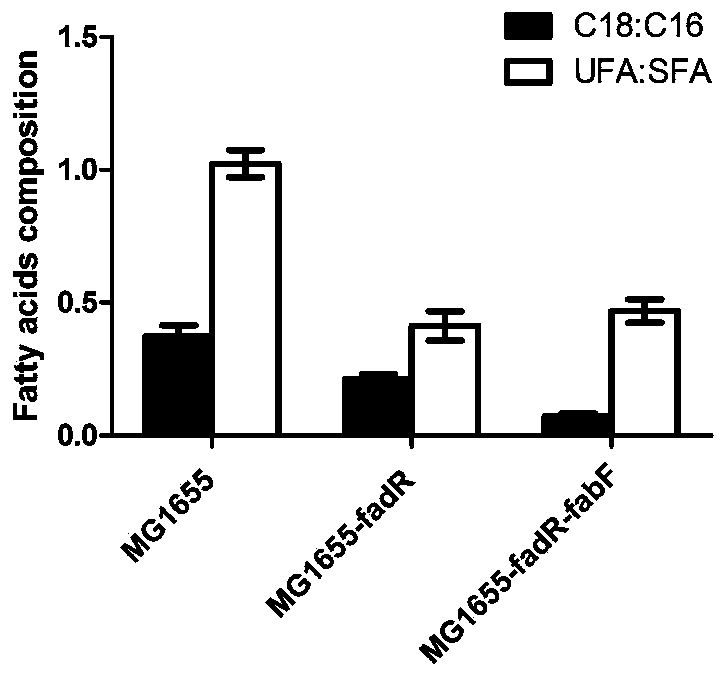
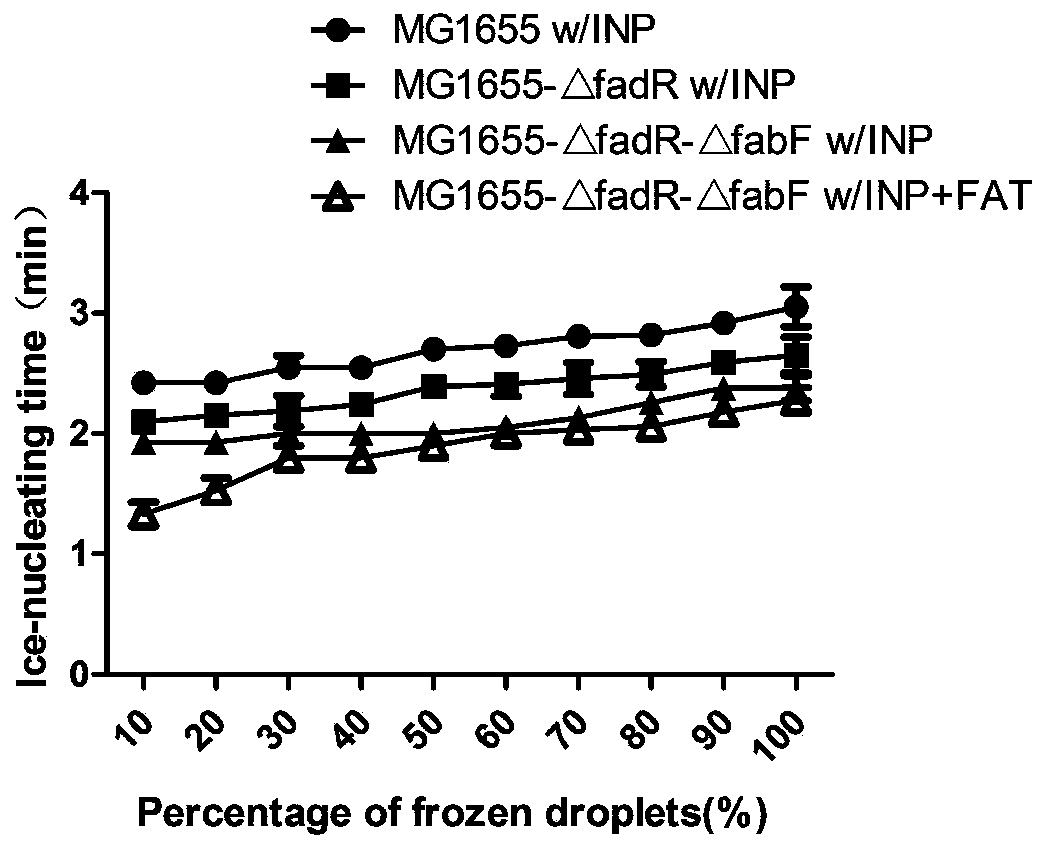
[0028] The inventors have found that the activity of the ice nucleation protein of the ice nucleation protein strain is related to the degree of fatty acid saturation and the average chain length of the bacterial strain. , in one of the most preferred schemes, compare the chain length and degree of saturation of three strains of wild-type Escherichia coli MG1655, DH5α, and BW25113, an...
Embodiment 2
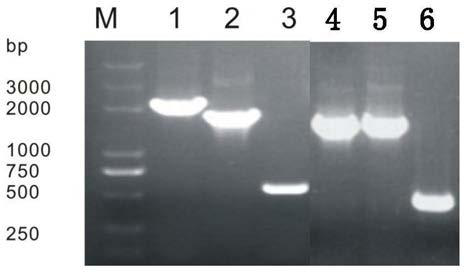
[0038] Example 2 Construction of MG1655-△fadR-△fabF Double Knockout Mutant
[0039] In order to obtain fatty acids with shorter chain length and higher degree of saturation, two genes in MG1655, β-ketoacyl-ACP synthase Ⅰ gene (FabF) and fatty acid degradation inhibitor gene (FadR), were knocked out, in which β- The nucleotide sequence of the ketoacyl-ACP synthetase I gene (FabF) is SEQ ID NO.1, and the nucleotide sequence of the fatty acid degradation inhibitor gene (FadR) is SEQ ID NO.2.
[0040] SEQ ID NO.1
[0041] ATGTTGTCTCCTGTCGGCAATACCGTAGAGTCTACCTGGAAAGCTCTGCTTGCCGGTCAGAGTGGCATCAGCCTAATCGACCATTTCGATACTAGCGCCTATGCAACGAAATTTGCTGGCTTAGTAAAGGATTTTAACTGTGAGGACATTATCTCGCGCAAAGAACAGCGCAAGATGGATGCCTTCATTCAATATGGAATTGTCGCTGGCGTTCAGGCCATGCAGGATTCTGGCCTTGAAATAACGGAAGAGAACGCAACCCGCATTGGTGCCGCAATTGGCTCCGGGATTGGCGGCCTCGGACTGATCGAAGAAAACCACACATCTCTGATGAACGGTGGTCCACGTAAGATCAGCCCATTCTTCGTTCCGTCAACGATTGTGAACATGGTGGCAGGTCATCTGACTATCATGTATGGCCTGCGTGGCCCGAGCATCTCTATCGCGACTGCCTGTACTTCCGGCGT...
Embodiment 3
[0067] Example 3 Transfer of thioesterase gene into MG1655-△fadR-△fabF double knockout mutant
[0068] Using the genome of Arabidopsis thaliana as a template to clone the thioesterase gene AtFat, its nucleotide sequence is shown in SEQ ID NO:3:
[0069] ATGTGACTGATTCTAAGTTACAGAGAAGCTTACTCTTCTTCTCCCATTCATATCGATCTGATCCGGTGAATTTCATCCGTCGGAGAATTGTCTCTTGTTCTCAGACGAAGAAGACAGGTTTGGTTCCTTTGCGTGCTGTTGTATCTGCTGATCAAGGAAGTGTGGTTCAAGGTTTGGCTACTCTCGCGGATCAGCTCCGATTAGGTAGTTTGACTGAAGATGGTTTATCTTATAAAGAGAAGTTTGTTGTTAGATCTTACGAAGTGGGTAGTAACAAAACCGCTACTGTTGAAACCATTGCTAATCTTTTACAGGAGGTGGGATGTAATCATGCACAAAGTGTTGGTTTTTCGACTGATGGGTTTGCAACAACAACTACTATGAGGAAGTTGCATCTCATTTGGGTTACTGCGAGAATGCATATCGAGATCTATAAGTACCCTGCTTGGGGTGATGTGGTTGAGATAGAGACTTGGTGTCAGAGTGAAGGAAGGATTGGGACAAGGCGTGATTGGATTCTTAAGGATTCTGTCACTGGTGAAGTCACTGGCCGTGCTACAAGCAAGTGGGTGATGATGAACCAAGACACGAGACGGCTTCAGAAAGTTTCTGATGATGTTCGGGACGAGTACTTGGTCTTCTGTCCTCAAGAACCGAGGTTAGCATTTCCGGAAGAGAATAACAGAAGCTTGAAGAAAATCCCGAAACTCGAAGATCCGGCTCAGTATTCAATGATTGG...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com