Evaluation method and system of anti-salt property of copolymer based on molecular simulation
A molecular simulation and evaluation method technology, applied in the fields of instrumentation, computational theoretical chemistry, informatics, etc., can solve problems such as poor repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
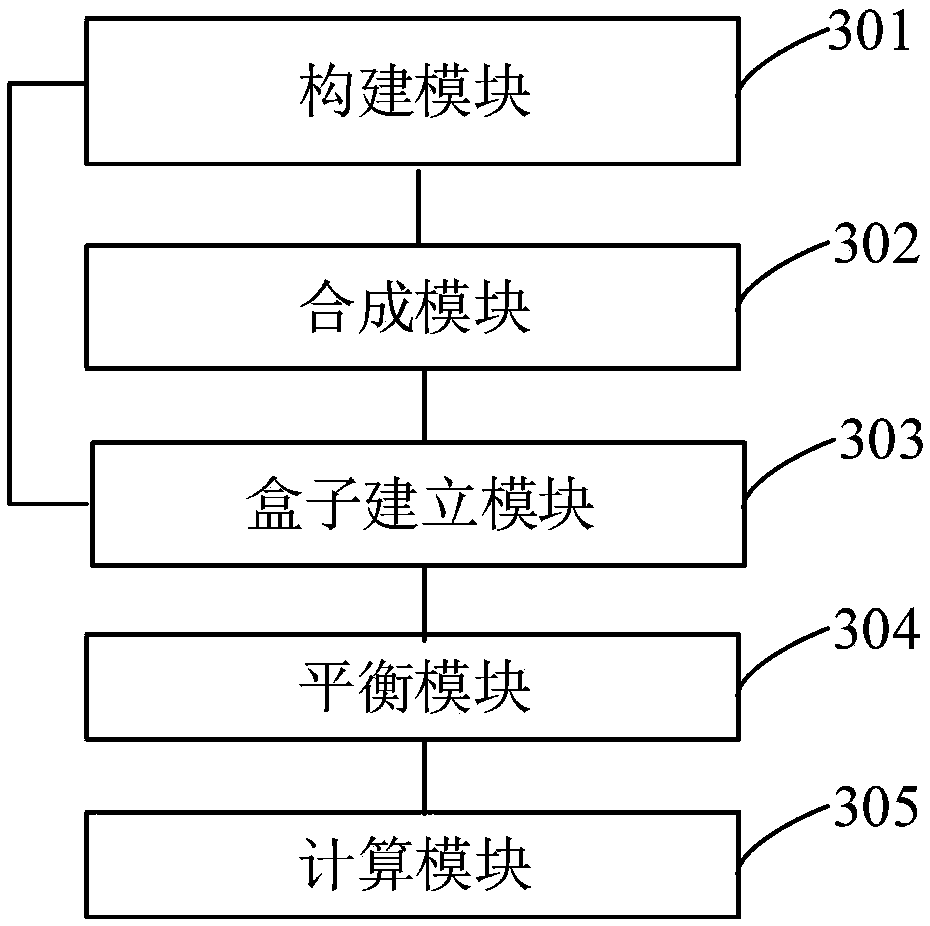
Method used
Image
Examples
example 1
[0085] Example 1: Evaluation of the salt resistance of copolymer P (AM-AA) at 1% NaCl ion concentration
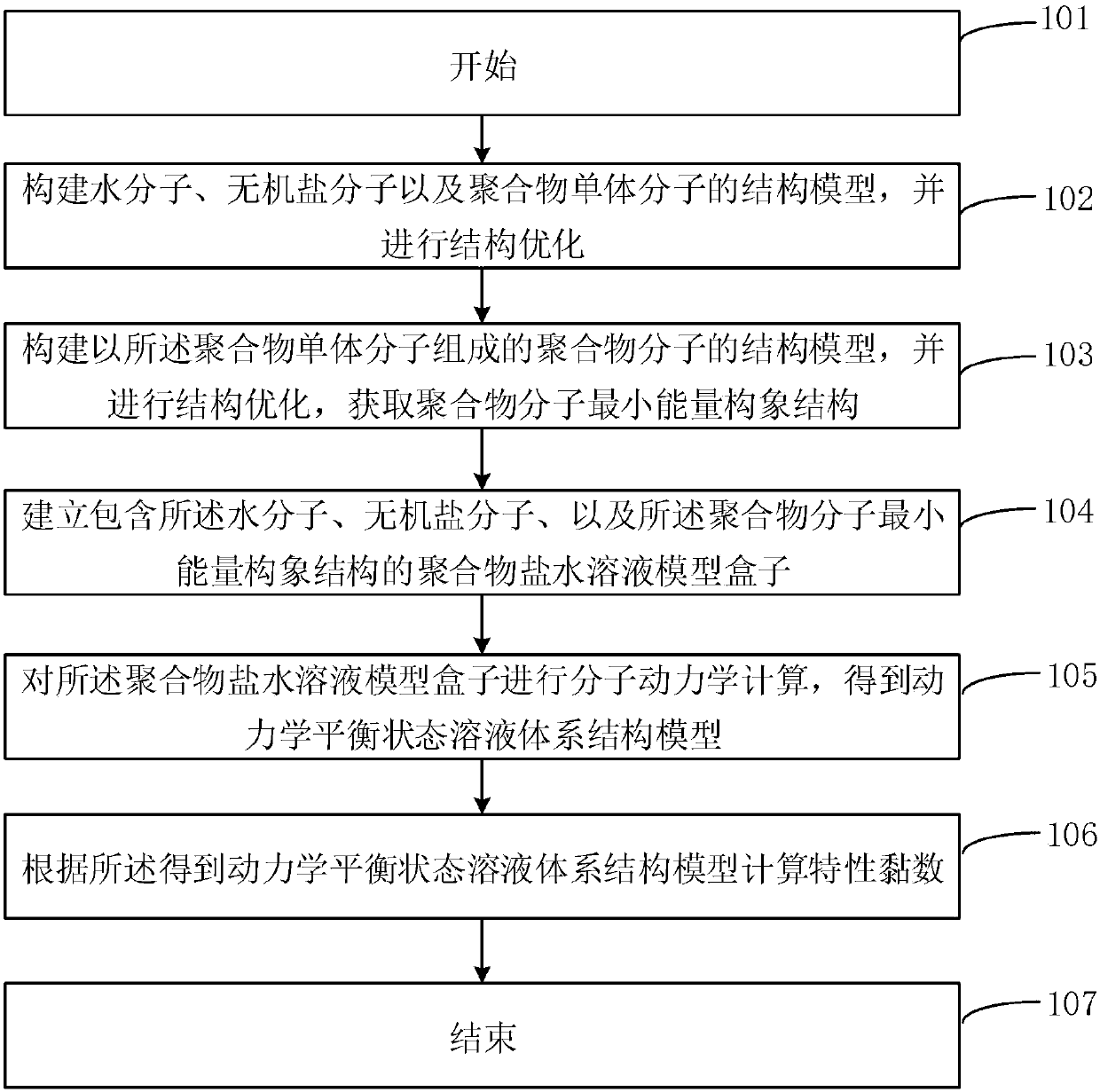
[0086] In step 102, the molecular models of water molecules, NaCl molecules, polymer monomers polyacrylamide (AM) and polyacrylic acid (AA) are drawn with the drawing tool of the Materials Studio software, and the structure is optimized through the Forcite module, using the COMPASSII force field.
[0087] In step 103, the random copolymer of AM and AA is constructed by the Biuld tool and is denoted as P(AM-AA), the AM and AA monomer models that have been built are selected, the input chain length is 10, and the input monomer molar ratio is 0.5 :0.5, establish P(AM-AA), and optimize the structure through Forcite.
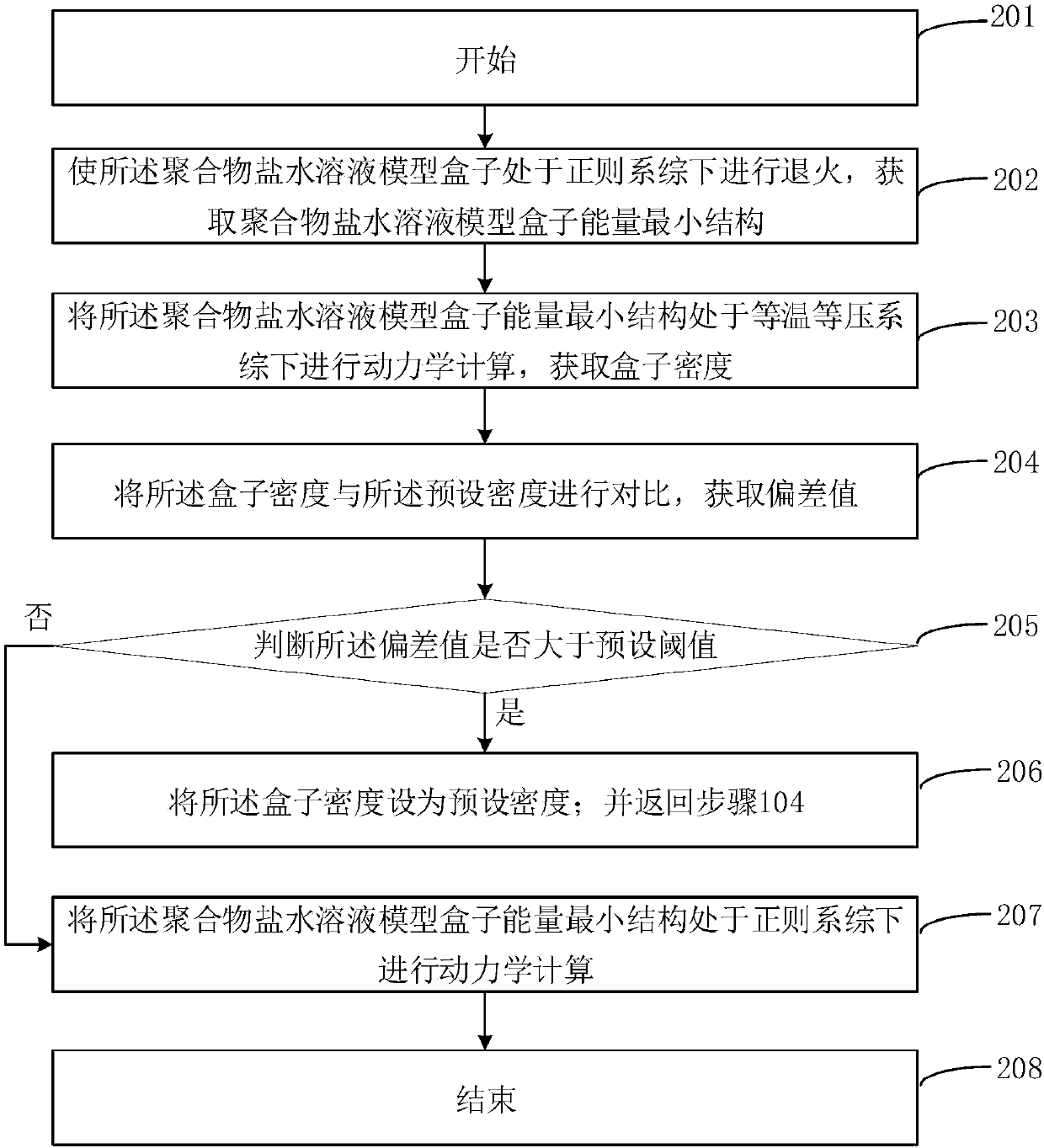
[0088] In step 104, the polymer solution model is established through the Amorphous Cell module, and the system density is set to 1.0g / cm 3 , select P(AM-AA), set the number of molecules to 1, select NaCl molecules, set the number of molecules to 3, select water...
example 2
[0091] Example two: evaluation of copolymer P (AM-AA-AMPS) salt resistance performance at 1% NaCl ion concentration
[0092] In step 102, the water molecule, NaCl molecule, polymer monomer AM, AA, 2-acrylamide-2-methylpropanesulfonic acid (AMPS) molecular model is drawn by the Materials Studio software drawing tool, and the structure is carried out by the Forcite module Optimization, using COMPASSII force field.
[0093] In step 103, build the random copolymer of AM, AA, AMPS by Biuld tool and be denoted as P (AM-AA-AMPS), select the AM, AA, AMPS monomer model that has built, input chain length 10, input The monomer molar ratio is 0.5:0.25:0.25, P(AM-AA-AMPS) is established, and the structure is optimized by Forcite.
[0094] In step 104, the polymer solution model is established through the Amorphous Cell module, and the system density is set to 1.0g / cm 3 , select P(AM-AA-AMPS), set the number of molecules to 1, select NaCl molecules, set the number of molecules to 6, selec...
example 3
[0097] Example three: evaluation of copolymer P (AM-AA-AMPS) in 1% CaCl 2 Salt resistance under ion concentration
[0098] In step 102, draw water molecule, CaCl by Materials Studio software drawing tool 2 Molecule, polymer monomer AM, AA, AMPS molecular model, structure optimization through Forcite module, using COMPASSII force field.
[0099] In step 103, build the random copolymer of AM, AA, AMPS by Biuld tool and be denoted as P (AM-AA-AMPS), select the AM, AA, AMPS monomer model that has built, input chain length 10, input The monomer molar ratio is 0.5:0.25:0.25, P(AM-AA-AMPS) is established, and the structure is optimized by Forcite.
[0100] In step 104, the polymer solution model is established through the Amorphous Cell module, and the system density is set to 1.0g / cm 3 , select P(AM-AA-AMPS), set the number of molecules to 1, and select CaCl 2 For molecules, set the number of molecules to 3, select water molecules, set the number of molecules to 2000, set the temp...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Density | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com