A kind of chcdc10 gene of Pseudomonas maize and its application
A technology of small spot bacterium and gene of corn, applied in the field of discovery and the application of its encoded protein, can solve problems such as loss
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1 Correlation analysis of the ChCDC10 gene of maize spot bacterium
[0038] The ChCDC10 gene of P. maize was obtained by comparison with the CDC gene in yeast by our team in P. maize. The open reading frame of ChCDC10 gene of maize leaf spot consists of 1197 nucleotides, including 2 introns. The encoded protein product consists of 341 amino acids, and domain analysis revealed that the ChCDC10 protein contains a conserved CDC-Septin domain (see figure 1 ).
Embodiment 2
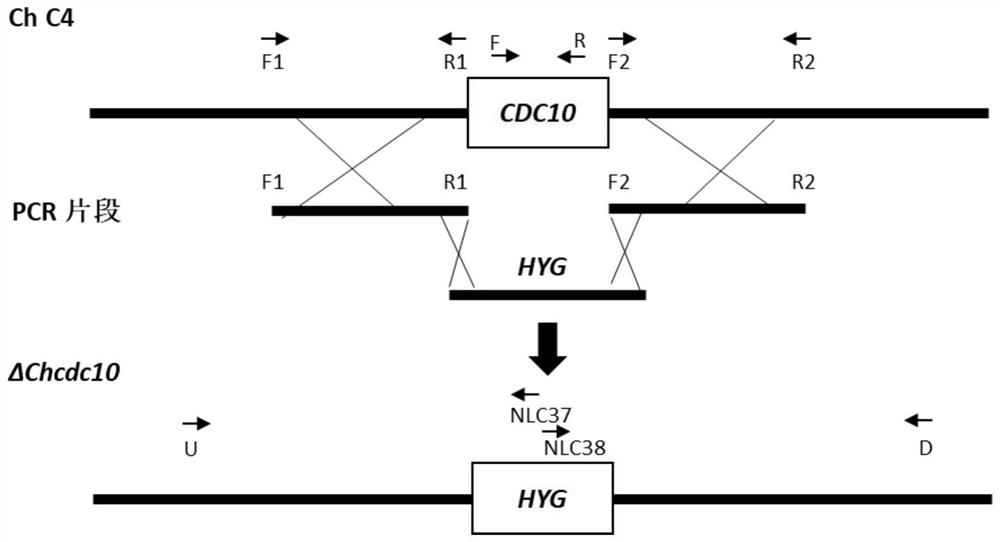
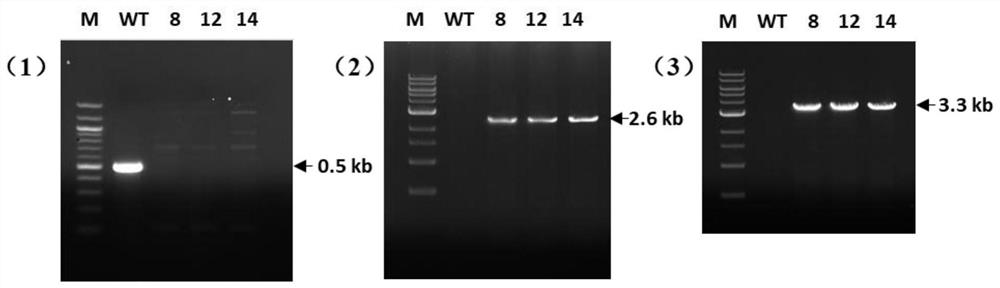
[0039] Example 2 Knockout of the ChCDC10 gene of P. maize spot bacterium
[0040] 1) Amplification of the upstream and downstream of ChCDC10 gene and hygromycin gene
[0041] Primers F1 (5'-GCCATTCCTACGTCAAAACC-3') and R1 (5'-TCCTGTGTGAAATTGTTATCCGCTGGCAGACGGACAAGGTAAAA-3') were used to amplify the upstream 888p fragment of ChCDC10 gene using F2(5 '-GTCGTGACTGGGAAAACCCTGGCGTCGCGTCGATAGCAATACAG-3') and R2 (5'-GACACGGCAAACACTGAAGA-3') to amplify the 884bp fragment downstream of the ChCDC10 gene of P. 3'), using the vector pUCATPH as a template to amplify the 2549bp hygromycin gene. The reaction system is: 10mmol / LdNTPMixture, 1μL; 5×PCRbuffer, 10μL; each 2.5μL of upstream and downstream primers (10μmol / mL); template DNA, 2μL; Phusionpolymerase, 0.5μL (5U); ddH 2 O, 31.5 μL; amplification program: 98°C pre-denaturation for 2 minutes, then (1) 98°C, denaturation for 20 seconds; (2) 65°C, annealing for 30 seconds; (3) 72°C, extension for 30 seconds; (4) ) cycled 30 times; (5) ex...
Embodiment 3
[0049] Example 3 Genetic Complementation of ChCDC10 Gene Deletion Mutant
[0050]Primers C-F1 (5'-GCTCTAGATGAGCTGACCGAAGATGTTG-3') and C-R1 (5'-CACTGGAACAACTGGCATGTTTGAGAAGTTTGCCGCTCT-3') were used to amplify the full-length 3112bp (including upstream and downstream sequences) of the ChCDC3 gene of P. F2 (5'-CAGGTACACTTGTTTAGAGGT CGTGTTGTTTCTCCAAGCTG-3') and C-R2 (5'-GGGCAGAATCTTCTTTGGTG-3') amplify the downstream sequence 419bp downstream of the ChCDC3 gene of B. maize. Then the vector pⅡ99 was used as a template, and the nptⅡ gene was amplified with DW69 (5'-CATGCCAGTTGTTCCAGTG-3') and DW70 (5'-ACCTCTAAACAAGTGTACCTG-3') primers. The three complementary fragments were transferred into the genome of the ChCDC10 gene deletion mutant, and Geneticin was used as a selection marker to screen the genetic complementation strain ΔChCDC10-C. Primers F / R were selected for PCR verification.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com