Transformant specific detection primer for transgenic insect-resistant and herbicide-resistant corn CM8101 and application thereof in backcrossing
A technology for herbicide tolerance and detection of primers, applied in the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc., can solve unrealistic problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Example 1 Integration of the target gene in the genome of insect-resistant and herbicide-resistant transgenic maize CM8101
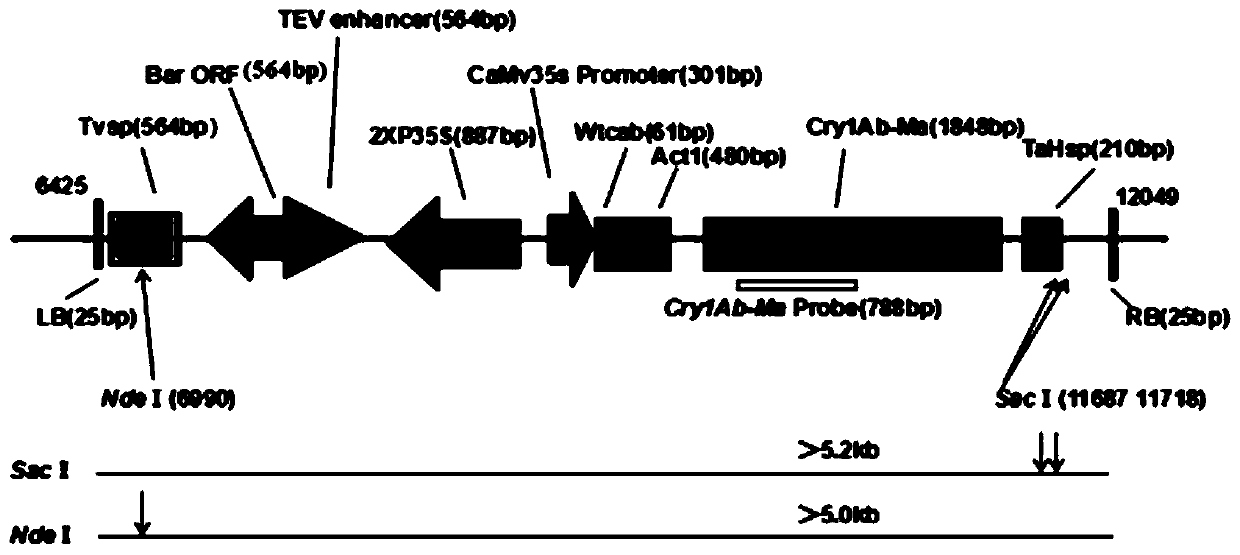
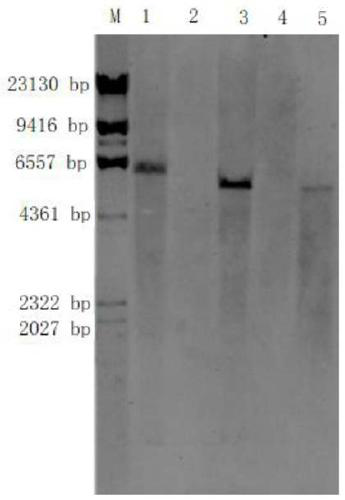
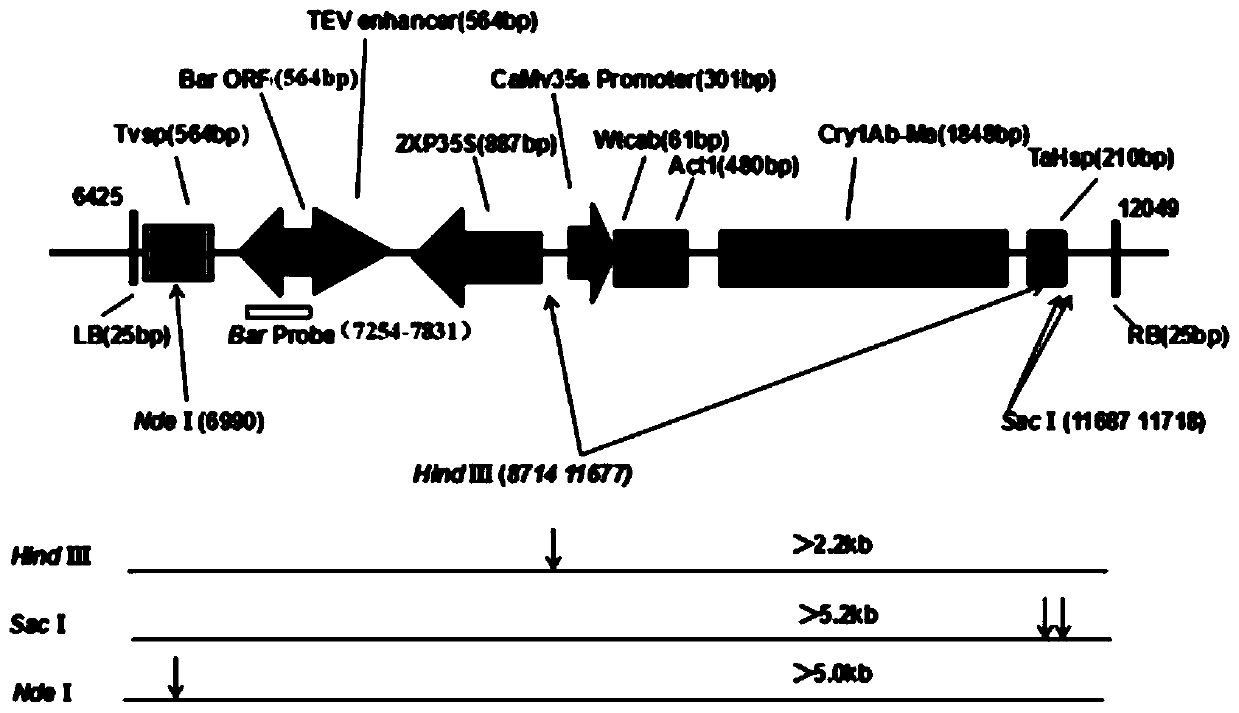
[0024] 1. Southern blot hybridization detection technology
[0025] (1) Agarose electrophoresis: Take a certain amount of DNA sample to be tested, and digest 10-20 μg with an appropriate restriction endonuclease; after digestion, perform electrophoresis on agarose gel; after electrophoresis, transfer to membrane.
[0026] (2) Blot transfer: cut the gel and mark it (excision in the lower left corner) for easy positioning, then place the gel in a container, depurinate it with 0.25M HCl for 10 min; soak the gel in an appropriate amount of denaturing solution, Place it at room temperature for 30 minutes to denature it, and shake gently without interruption; cut the filter paper and nylon membrane to the same size as the glue, and immerse the nylon membrane in the transfer buffer for 30 minutes to balance; spread layer by layer, leaving no air bubbles ...
Embodiment 2
[0042] Example 2 Analysis of exogenous insertion sequence
[0043] The flanking genome sequence of the inserted fragment and the sequence inserted into the maize genome near the 5' end and 3' end were obtained by optimized Tail-PCR method.
[0044] According to the hiTAIL-PCR method reported by Liu Yaoguang et al., the specific primers for hiTAIL-PCR were designed according to the sequence of the 5' end and 3' end of the vector. The nucleotide sequence and the length of the amplified fragment are shown in Table 3. See Table 4 for arbitrary degenerate primers AD1-n (n is 1 to 4), AC-1 and AD6 used in hiTAIL-PCR.
[0045] Table 3 specific primers
[0046] serial number name Primer sequence (5'-3') SEQ ID No.1 LB0-1 AGGGGTGTGGAAATAGTGATTTGAGCACGTC SEQ ID No.2 LB1-1 ACGATGGACTCCAGTCCAACGTCGTGACTGGGAAAACCCTGGC SEQ ID No.3 LB2-1 TTGCAGCATCACCCCCTTTCGCCAGCT SEQ ID No.4 RB0-1 CTCAGGATTTAGCAGCATTCCGGGTAC SEQ ID No.5 RB1-2 GTAA...
Embodiment 3
[0065] Example 3 Backcross Transgenic Population Cry1Ab-Ma Gene-specific Qualitative PCR Detection
[0066] During the backcross breeding process, the CM8101 transformant-specific qualitative PCR was used to detect the primer 8101-5'F / R, and the prospects of the Cry1Ab-Ma gene were detected on the plants screened by glufosinate-ammonium and Bt test strips. The Cry1Ab-Ma gene was carried in the single plant used for backcrossing. The molecular marker 8101-5'F / R could amplify a 629bp fragment in the three generations (BC3, BC4, BC5) and CM8101 (T9) of nine backcross populations ( Figure 7 ), proving that the Cry1Ab-Ma gene in CM8101 was successfully transferred to 9 recipient inbred lines by backcrossing, and it was stably inherited in each backcrossing generation.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com