Preparation method of DNA nanowire cluster loaded with phenanthroline ruthenium
A technology of nanowires and ruthenium pyroxoline, applied in the field of analytical chemistry, can solve the problems of few biosensing applications, and achieve the effects of good reproducibility, simple synthesis method, and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach
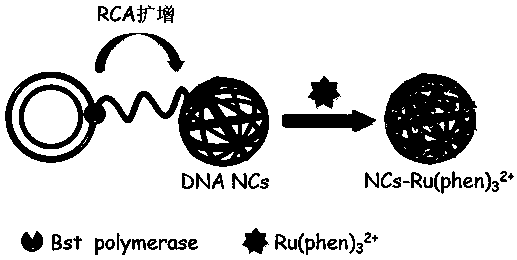
[0029] 1. Design and synthesize the palindromic sequence-encoded rolling circle amplification (RCA) reaction template strand and the corresponding primer strand. RCA amplification template strand sequence from left to right 5' to 3': phosphorylation-GGGCACGGGCAGCTTGCCGGTGGAAGCTAGATGCATCTAGCAAGCGCCGCCACTGATTTCACCGCTTCAAGCTAGATGCATCTAGCAAT; RCA amplification The primer strand sequence is from left to right 5' to 3': GTGGCGGCGC. .
[0030] 2. The ligation and cyclization process of the RCA template strands, the specific steps are as follows: 40 μL of the solution contains RCA template strands with a final concentration of 0.50 μM, 5.0 U μL -1 CircLigase II ssDNA Ligase, 2.5 mM Manganese Chloride (MnCl 2 ) and 1 M betaine, and incubated at 60 °C for 1 hour to form a circularized template strand.
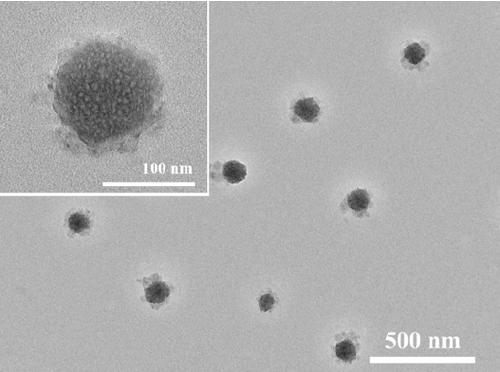
[0031] 3. Start the RCA amplification reaction to self-assemble and synthesize three-dimensional spherical DNA nanowire clusters (DNA NCs). The specific steps are as follows: add 2 μL...
Embodiment 1
[0037] 1. Prepare different sizes of DNA NCs-Ru(phen) 3 2+ The operation process of ECL analysis, the specific steps are as follows:
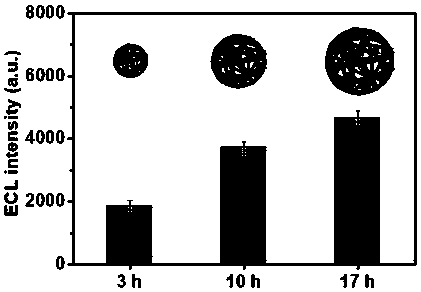
[0038] (1) DNA NCs of different sizes were synthesized by adjusting different RCA reaction times (3, 10, and 17 hours, respectively).
[0039] (2) The phenanthroline ruthenium (Ru(phen) 3 2+ ) loaded into the double-stranded structure of DNA NCs of different sizes to prepare DNANCs-Ru(phen) 3 2+ :
[0040] A: Take 10 uL DNA NCs and 1.0 uL phenanthroline ruthenium (Ru(phen) 3 2+ ) (concentration of 10 mM) was mixed and added to 380 μL buffer 1, and incubated at 4 °C for 3 hours;
[0041] B: Transfer the above solution to an ultrafiltration centrifuge tube (3 K), and centrifuge at 10700 rpm for 20 minutes in a refrigerated centrifuge;
[0042] C: Collect the prepared DNA NCs-Ru(phen) 3 2+ Beacons were stored in clean centrifuge tubes at 4°C until use.
[0043] (3) Take 10 uL of DNA NCs-Ru(phen) of different sizes prepared above 3 2+...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com