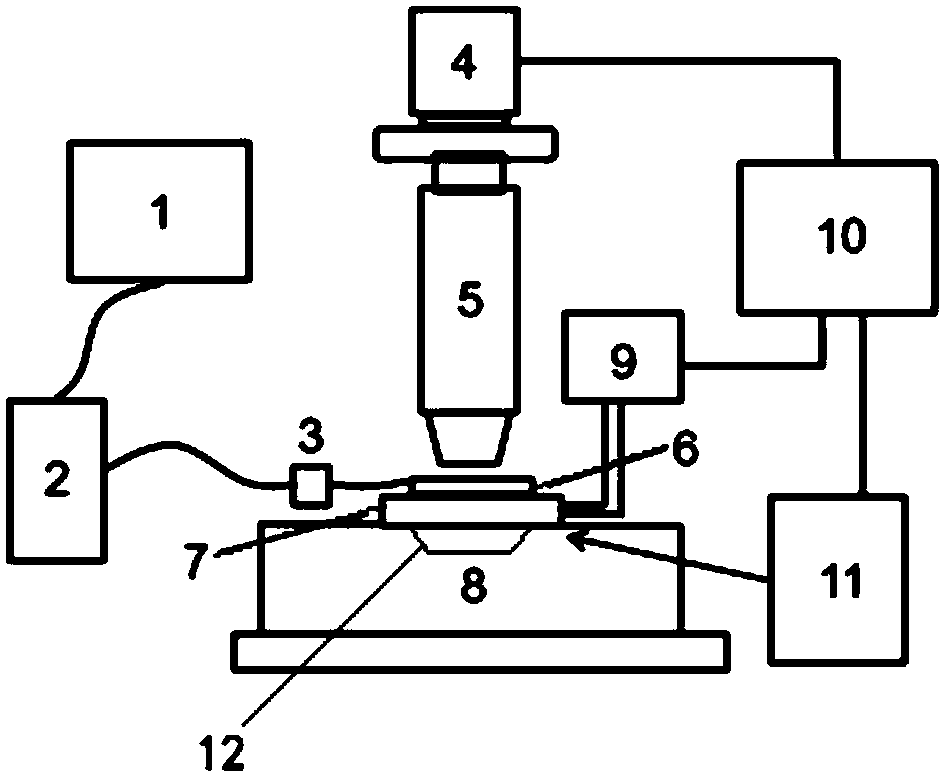
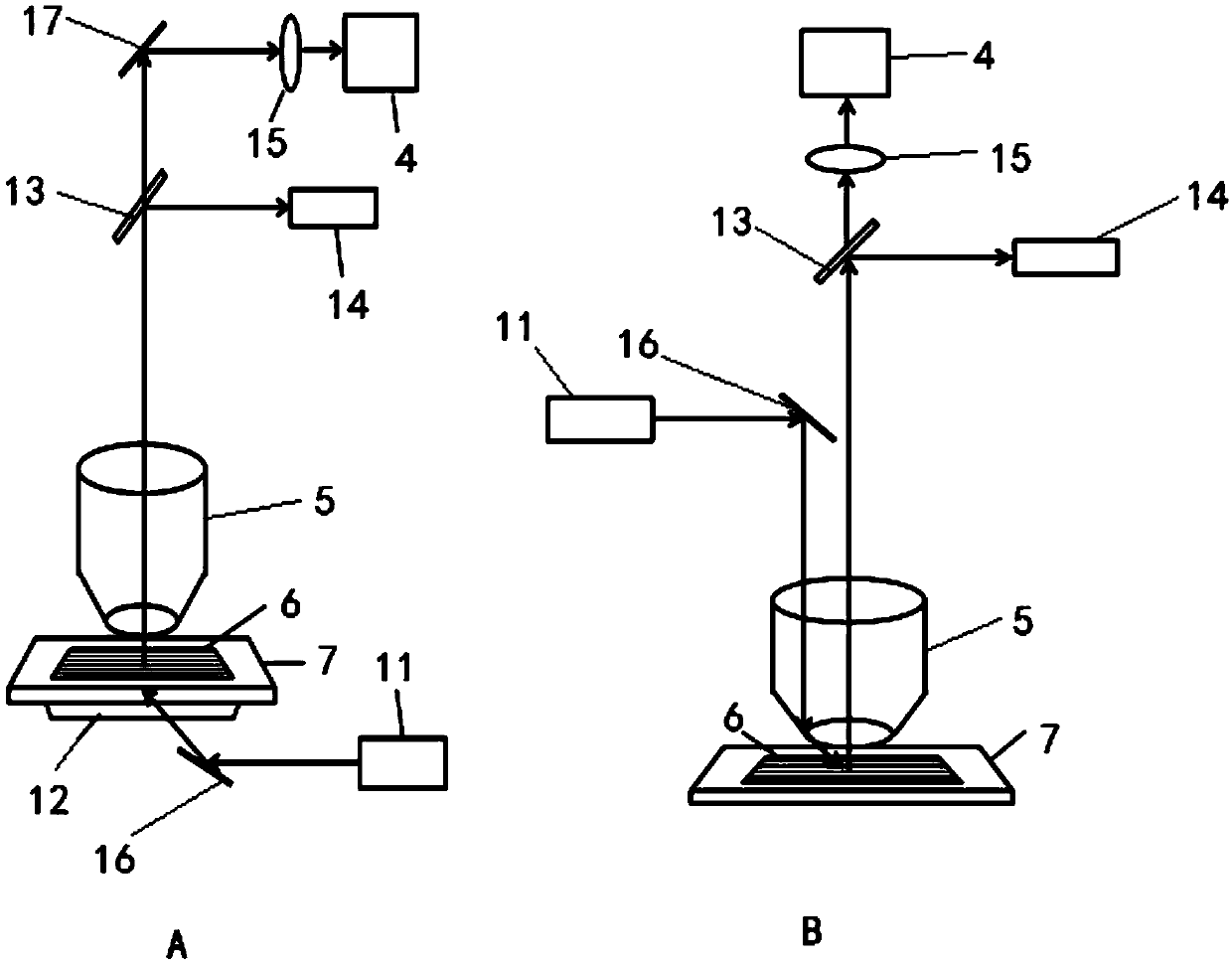
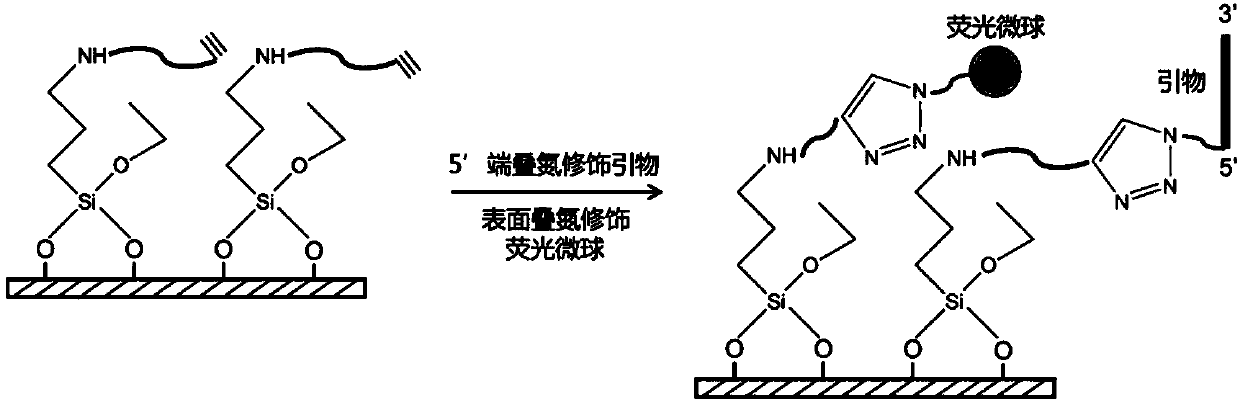
DNA single-molecule sequencing system and apparatus based on multicolor-fluorescence reversible termination nucleotide
A technique for terminating nucleotides and single-molecule sequencing, applied in the field of genetic engineering, can solve the problems of slow extension, fluorescence quenching, high mismatch rate, etc., and achieve the effect of precise focusing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0140] Example 1: Three-color fluorescent labeling reversible termination of nucleotide DNA single molecule sequencing system
[0141] The composition of the three-color fluorescent single-molecule sequencing system a1 in this embodiment is as follows: For the 3′-OH protected nucleotides without fluorescently labeled base G, it is composed of compound IV or VIII, and the corresponding fluorescently labeled base U nucleoside The acid is composed of one of compounds X, XIV, and XX; the corresponding fluorescently labeled base C nucleotide is composed of one of compounds XI, XVI, XXI, XXXVII, and the corresponding fluorescently labeled base A nucleotide is composed of XII, XVII, XXII, XXIII, XXXVIII; four modified nucleotides of different bases together form the sequencing reagent system 1 of this embodiment;
[0142] The composition of the reagent system a2 of the three-color fluorescent-labeled single-molecule sequencing system is as follows: For the 3′-OH protected nucleotides wit...
Embodiment 2
[0163] Example 2: Four-color fluorescent labeling reversible termination of nucleotide DNA single molecule sequencing system
[0164] In the four-color fluorescent single-molecule sequencing system of this embodiment, the reversible terminating nucleotide is selected in this embodiment. For base U, select XXVI, XXVII, XVIII, or XIX, for base C, select XXIX, for base G, XXXI or XXXII, select XXXV or XXXVI for base A, and four modified nucleotides of different bases together form the sequencing reagent system 1 of this embodiment;
[0165] Reagent system 2: compound X, XI, XII, XIII of the four-color fluorescent reversible termination nucleotide sequencing system of the present invention;
[0166] Reagent system 3: compound XIV, XVI, XVII, XV of the four-color fluorescent reversible termination nucleotide sequencing system of the present invention;
[0167] The four-color fluorescent reversible termination nucleotide sequencing system reagent system of the present invention 4: For U, s...
Embodiment 3
[0178] Example 3: Construction and application of single-molecule paired-end sequencing system
[0179] Use the sequencing chip of Example 1 to analyze the DNA template sequence to be tested (sequence 5)
[0180] 5'-GTTGTTGTTGTTGTTGTTCTACGTTCGAACTACTAAGCAATCCGGCAGATCGTCACAAAAAAAAAAAAAAAAAAAA-3' for double-ended single molecule sequencing. Specific steps are as follows:
[0181] (1) The primer immobilized on the surface of the substrate is 5'-(CNVC)TTUTTTTTTTTTTTTTTTTT-3' (sequence 6), where CNVC is a reversible light crosslinking agent between DNA strands, and then the template column to be sequenced is fixed with the The primers were incubated at 65°C for 5 minutes and slowly cooled to 37°C for the first hybridization; after the hybridization, four natural nucleotides (dATP, dTTP, dCTP, dGTP) were added, and the Under the action, the fixed primer is extended to synthesize a DNA strand complementary to the sequence of the template to be tested. Then the temperature was increased...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Thickness | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com