Use of biological RNA scaffolds with in vitro selection to generate robust small molecule binding aptamers for genetically encodable biosensors
A technology for combining structural domains and ligands, applied in the field of oligonucleotide libraries, can solve problems such as inability to integrate easily
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0194] Example 1: Construction of selection libraries using information from biological RNA
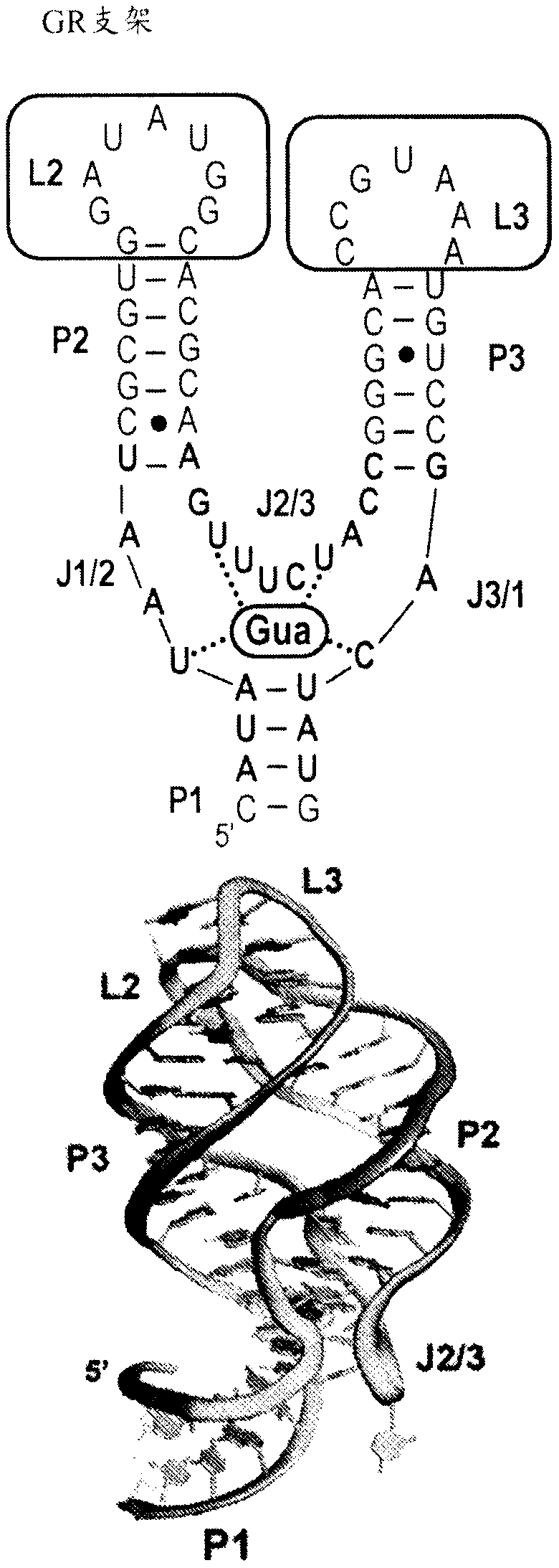
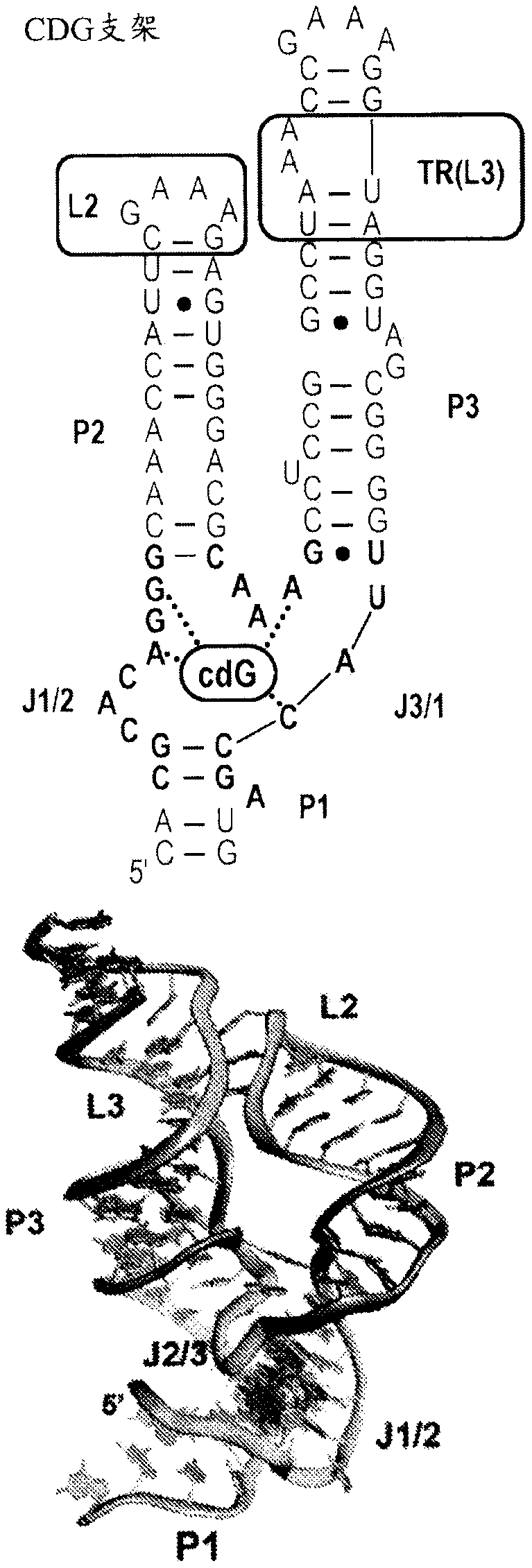
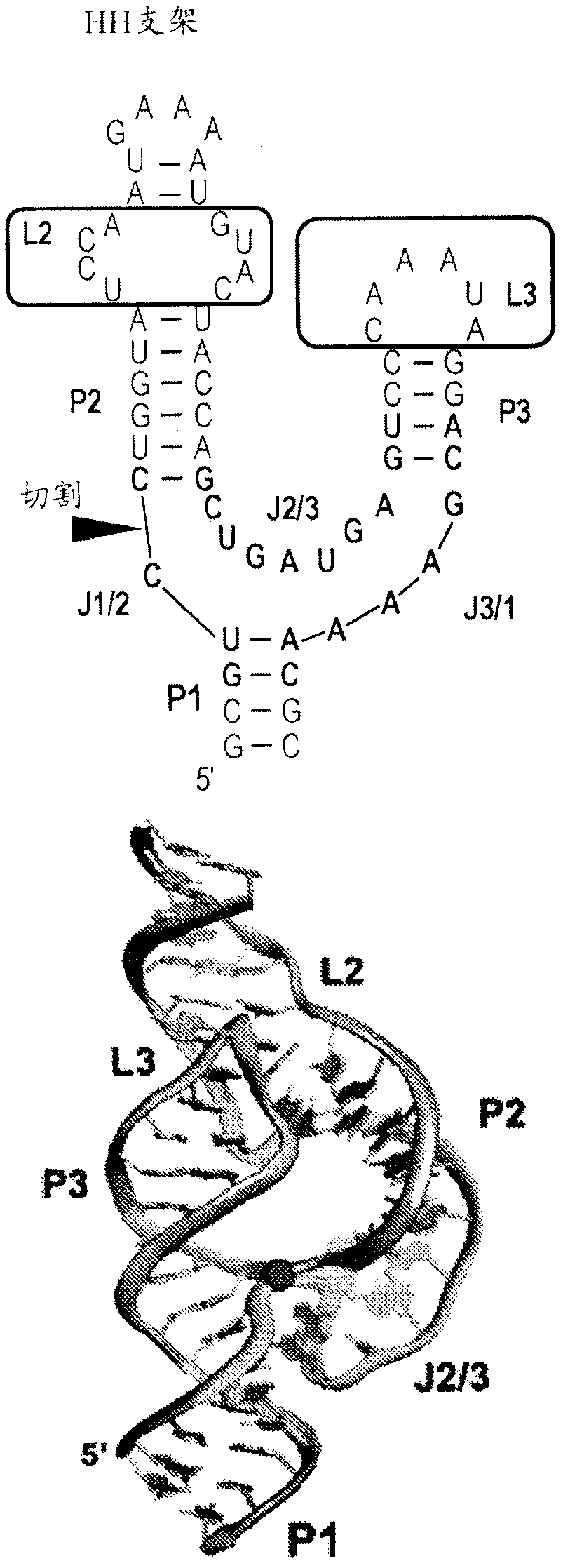
[0195] Examination of small biological RNAs with multihelical packing (ie, tertiary folding) indicated two recurring structures that could be considered privileged scaffolds. The first is the H-type pseudoknot, which is ubiquitous in biological RNAs, including small ribozyme ribosomal frameshift elements in viral mRNAs, and natural and synthetic aptamers. However, from a design point of view, such folding is difficult to design. Another is the three-way junction (3WJ), which is supported by long-range tertiary interactions that organize a helical arrangement around the junction. This fold is more suitable for the design of aptamer-incorporated RNA devices because it positions a designable helical element, called the P1 helix, near the ligand-binding site normally housed in the junction.
[0196] In the three-way junction foldome, there is a large number of potential candidate select...
Embodiment 2
[0208] Example 2: Scaffold selection for 5HTP yields many potential aptamers
[0209] The target of choice is 5-hydroxy-L-tryptophan (5HTP; Figure 2A ), the direct biosynthetic precursor of serotonin, which is immobilized on a solid substrate via its carboxylate group. Seven rounds of selection were performed on each library, with counterselection for L-tryptophan and increasingly stringent washing procedures in later rounds. In SSIII selection, a conventional SELEX protocol was employed in which the affinity column was washed extensively in early rounds before competitive elution to remove non-bound RNA. Competitive elution was initially observed in round four and peaked at >50% of total input RNA in round six. GsI selection uses a less stringent protocol than is generally recommended, where approximately the final 10% of total RNA remaining on the column under competitive elution is collected for amplification in the first four rounds to increase wash stringency. Maintai...
Embodiment 3
[0218] Example 3: The densest clusters retain the scaffold structure and bind 5HTP with high selectivity
[0219] Structural scaffolds greatly facilitate the verification of the structural and interaction features of the resulting aptamers. Chemical probing of RNA structures using N-methylisatoic anhydride ("NMIA") (a technique known as "SHAPE") reveals the secondary and tertiary structure of the parental scaffold as well as ligand-dependent binding in the aptamer. Whether structural changes are preserved. In GR / SSIII selection, the NMIA reactivity patterns of the 5HTP-I and 5HTP-II aptamers changed locally in the presence of ligand within the three-way junction element, consistent with this being the ligand-binding site ( Figure 3A , Figure 19 ). However, 5HTP-III displayed changes outside of J2 / 3 in the constant region, consistent with the predicted structure of a previously described tryptophan aptamer and L-Trp binding site (Majerfeld and Yarus). Retention of the GR ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com