A method for detecting bacterial drug resistance based on AIE fluorescent probe
A fluorescent probe and fluorescence detection technology, which is applied in the direction of fluorescence/phosphorescence, measuring devices, and material analysis through optical means, can solve the problems of strong subjectivity in result judgment, complicated operation, and high repeatability, and achieve convenient and fast detection process , High detection sensitivity, easy to achieve effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
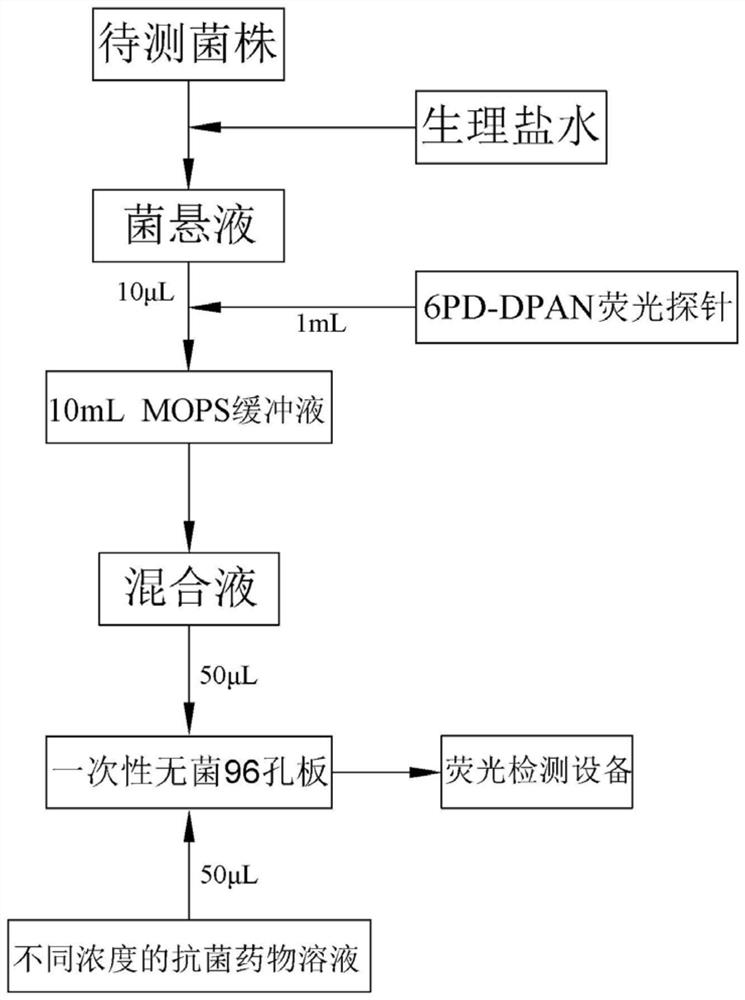
[0047] See figure 1 , 4 , a method for detecting bacterial drug resistance based on AIE fluorescent probes, comprising the steps of:
[0048] Step (1): Prepare the Escherichia coli strain with physiological saline to a bacterial suspension with a McFarland turbidity of 0.5;
[0049] Step (2): Dilute 1mL of 6PD-DPAN fluorescent probe and 10μL of the bacterial suspension in step (1) into 10mL of MOPS buffer, shake well to prepare a mixture;
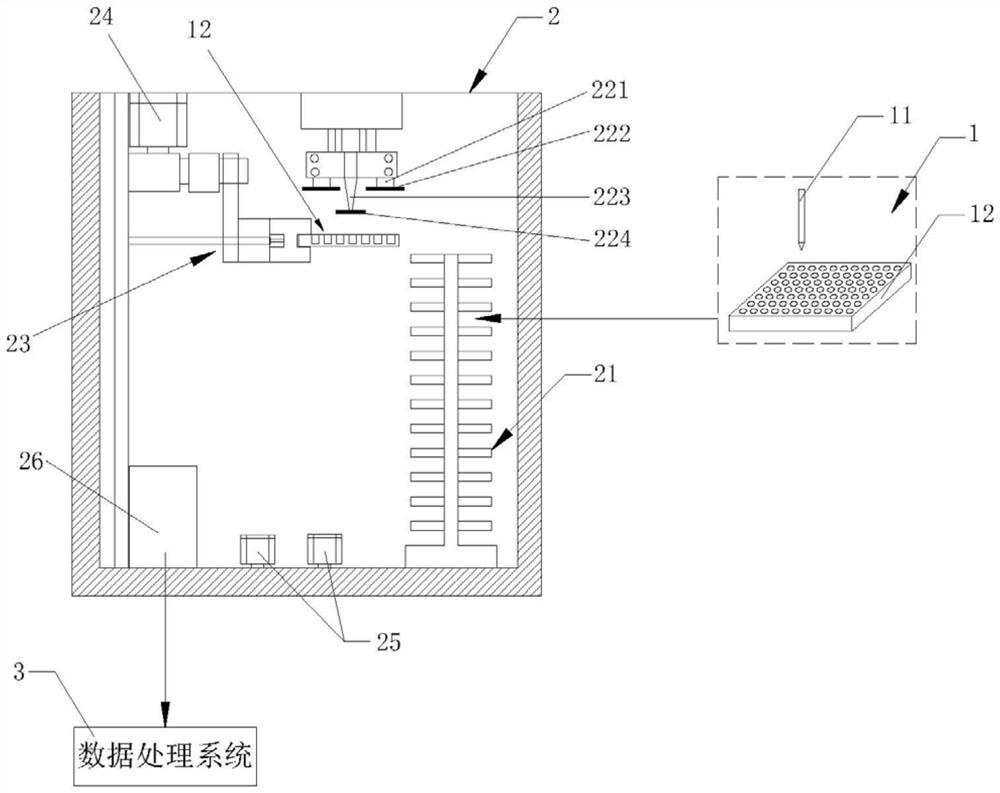
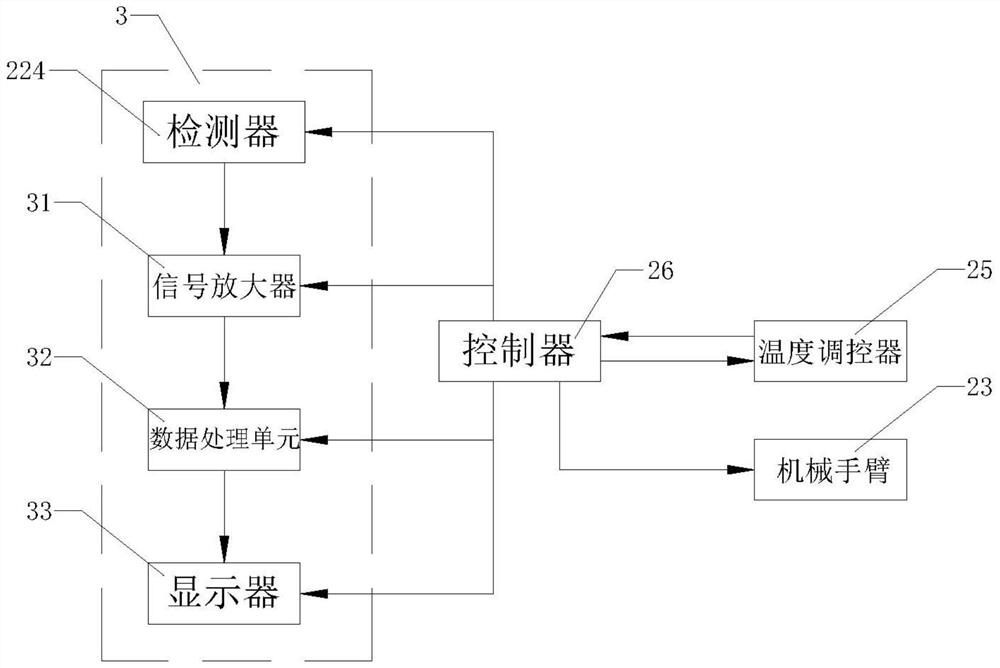
[0050] Step (3): Add 50 μL of the mixed solution prepared in step (2) to each well of the disposable sterile 96-well plate 11, and then add it to the first well of the disposable sterile 96-well plate 11 50 μL of MOPS buffer solution was prepared as a control sample, and 50 μL of antibacterial drug solutions of different concentrations were added to each well except the first well to prepare several samples to be tested, and the plate was sealed with a transparent film to detect and Record the initial fluorescence intensity value FRET of...
Embodiment 2
[0065] See figure 1 , 5 , a method for detecting bacterial drug resistance based on AIE fluorescent probes, comprising the steps of:
[0066] Step (1): The Klebsiella pneumoniae strain is prepared with physiological saline to a bacterial suspension with a McFarland turbidity of 0.5;
[0067] Step (2): Dilute 1mL of 6PD-DPAN fluorescent probe and 10μL of the bacterial suspension in step (1) into 10mL of MOPS buffer, shake well to prepare a mixture;
[0068] Step (3): Add 50 μL of the mixed solution prepared in step (2) to each well of the disposable sterile 96-well plate 11, and then add it to the first well of the disposable sterile 96-well plate 11 50 μL of MOPS buffer solution was prepared as a control sample, and 50 μL of antibacterial drug solutions of different concentrations were added to each well except the first well to prepare several samples to be tested, and the plate was sealed with a transparent film to detect and Record the initial fluorescence intensity valu...
Embodiment 3
[0082] See figure 1 , 6 , a method for detecting bacterial drug resistance based on AIE fluorescent probes, comprising the steps of:
[0083] Step (1): Prepare the Staphylococcus aureus strain with physiological saline to a bacterial suspension with a McFarland turbidity of 0.5;
[0084] Step (2): Dilute 1mL of 6PD-DPAN fluorescent probe and 10μL of the bacterial suspension in step (1) into 10mL of MOPS buffer, shake well to prepare a mixture;
[0085] Step (3): Add 50 μL of the mixed solution prepared in step (2) to each well of the disposable sterile 96-well plate 11, and then add it to the first well of the disposable sterile 96-well plate 11 50 μL of MOPS buffer solution was prepared as a control sample, and 50 μL of antibacterial drug solutions of different concentrations were added to each well except the first well to prepare several samples to be tested, and the plate was sealed with a transparent film to detect and Record the initial fluorescence intensity value FR...
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com