A highly efficient method for detecting protein subcellular localization based on citrus protoplasts
A protoplast, protein technology, applied in biochemical equipment and methods, microbial determination/inspection, biological testing, etc., can solve the problems of low success rate, high cost, complicated process, etc. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
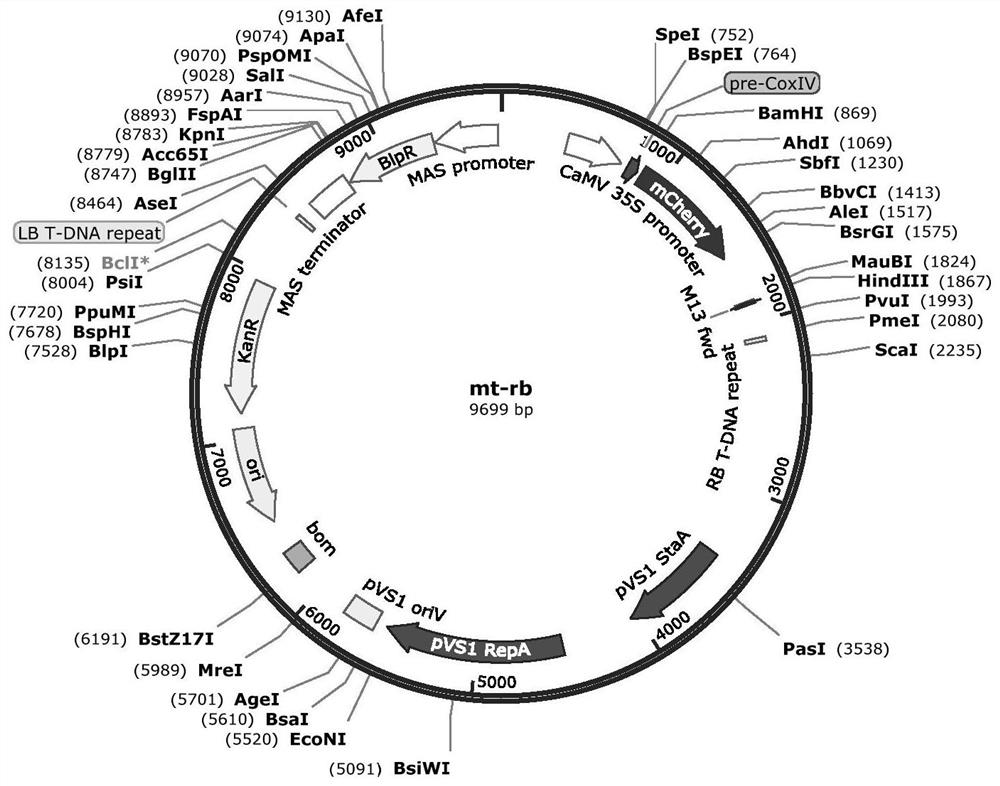
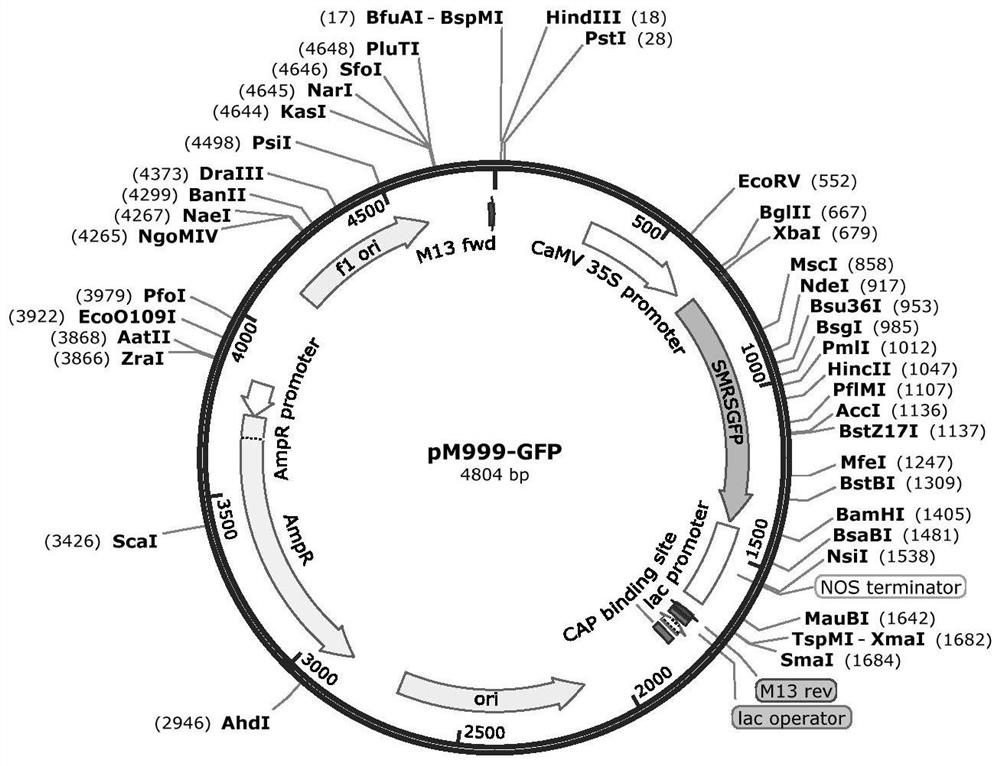
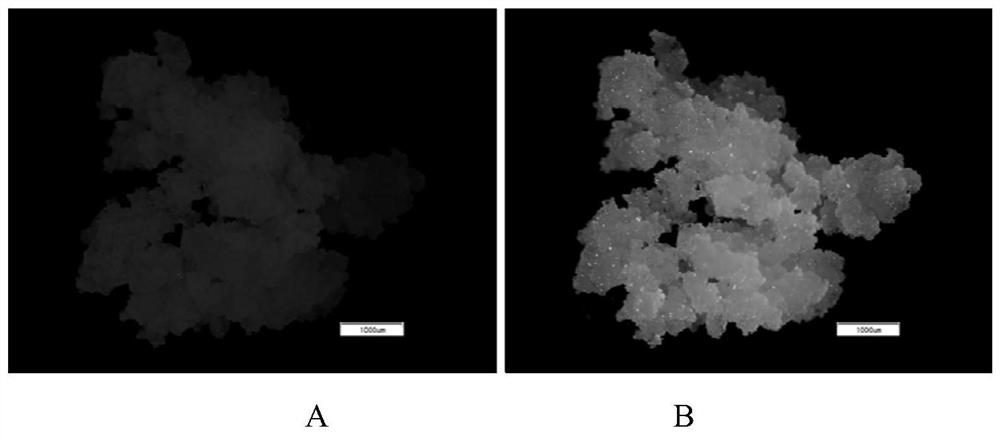
[0032] This example relates to the stable genetic transformation of citrus callus and the protoplast transformation of citrus callus. The callus stable genetic transformation vector is a fusion vector carrying mitochondrial-localized red fluorescent protein RFP (Red fluorescent protein) (the mitochondrial localization tag is fused with red fluorescent protein, and the expression is driven by the same promoter), and the vector map is detailed in figure 1, the carrier resistance is herbicide Basta resistance, denoted as mt-rb (Nelson B K, Cai X, Andreas Nebenführ. A multi-color set of in vivo organelle markers for colocalization studies in Arabidopsis and other plants[J]. The Plant Journal , 2007, 51(6):1126-1136.). The cloned coxII expression product of the gene to be tested is located on the mitochondria. The green fluorescent marker GFP (Green fluorescent protein) expression subcellular localization vector pM999-GFP used comes from the rice research team of the State Key Labo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com